The Autumn Upgrade: Minimap2 integration, SVG output and other features
Minimap2
Our next major feature upgrade of Persephone includes interesting new functions. First of all, we integrated minimap2 – a popular alignment tool that, besides aligning short reads, can be used for sequence comparisons on the scale of full chromosomes.
As you probably know, our popular “instant BLAST” feature aligns two sequences of the size up to 1 Mbp with a single-base precision. This tool will not work if you want to compare bigger regions. The introduction of minimap2 helps overcome this limitation. The users can now display two chromosomes side by side and push a button that will extract the genomic sequences and pass them to minimap2. In a couple of seconds, Persephone will graphically display the minimap2 output similar to this:
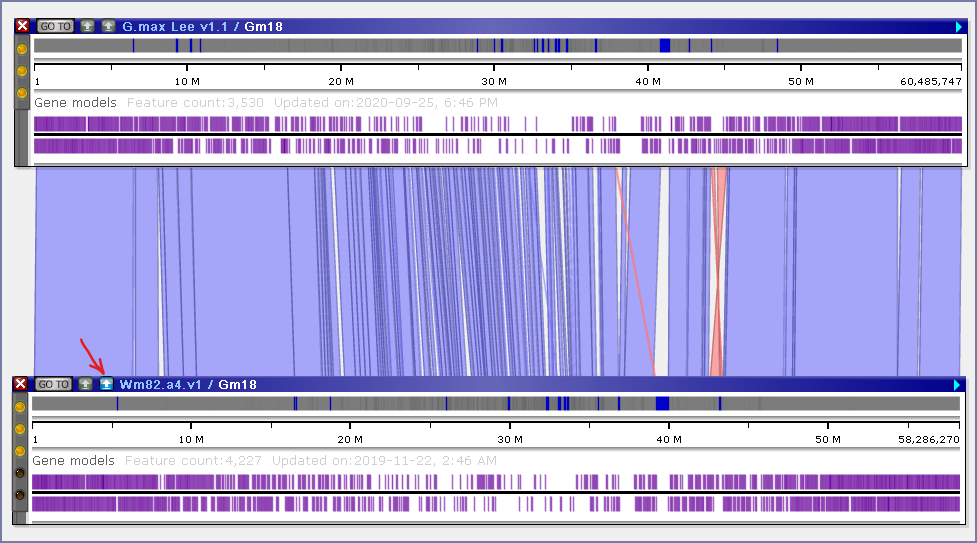
The blue and orange ribbon-like elements correspond to the “chains” produced by minimap2 that link regions of high sequence similarity. This real-time alignment will also work with the sequences from the user storage, typically added via drag and drop. This is quite valuable as it allows producing new map alignments right away, without any extra processing like mapping common markers or finding orthologous gene pairs.
Please note that you can align entire chromosomes if their combined length does not exceed 200 Mbp. In case of larger sequences, a message will be displayed warning about a possible time-out. Adjust the minimap2 parameters if necessary.
Launch Persephone and open an external file referenced via a URL
If you have a data track in the form of an external file, you can open it and view the data as a new track on a map. The pointer to the file should be embedded in the URL that starts Persephone using the argument ‘files=‘, for example,
You will also need to tell which genome the new track will be added to.
Launch Persephone and jump to a gene or marker specified in the URL
An annotation feature or a marker name can be specified in the URL that launches Persephone. Use the argument searchAnnot= or searchMarker= with the keyword. If the feature is found, Persephone will open a proper map and will zoom to the corresponding location. If the feature name produces multiple hits, the search results form will be shown.
Different shading shows the original exons in a spliced sequence.
Using slightly different backgrounds will help visualize the history of the spliced sequence showing individual exons that were spliced together. It works in the protein sequence tab too.

Protein tab 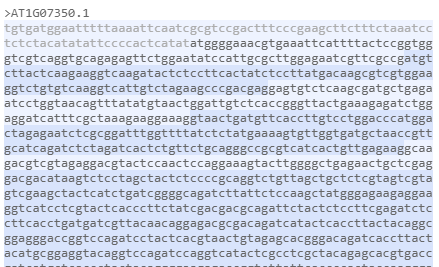
Spliced sequence tab
Synchronized sequence selection in different tabs
You can now select a sequence in one tab of the annotation details form, for example, the spliced sequence, and see which amino acids correspond to the selected DNA sequence in the protein or translation tab. Note that selecting a sequence of an exon or a UTR can be done by double-clicking in the corresponding section in the text.
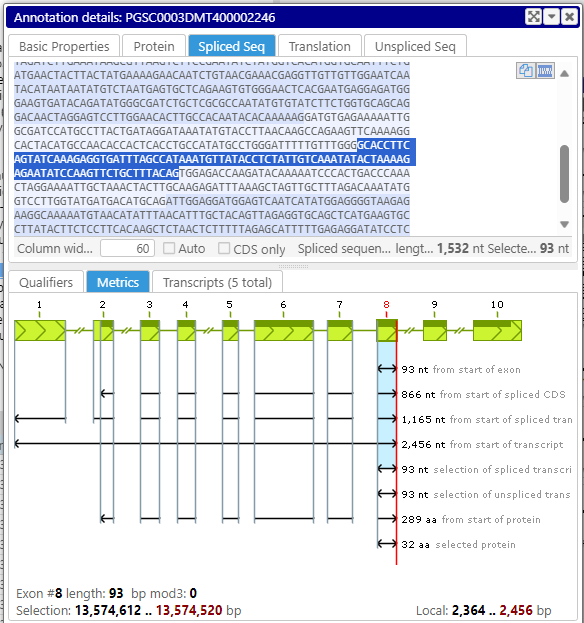
Spliced sequence tab 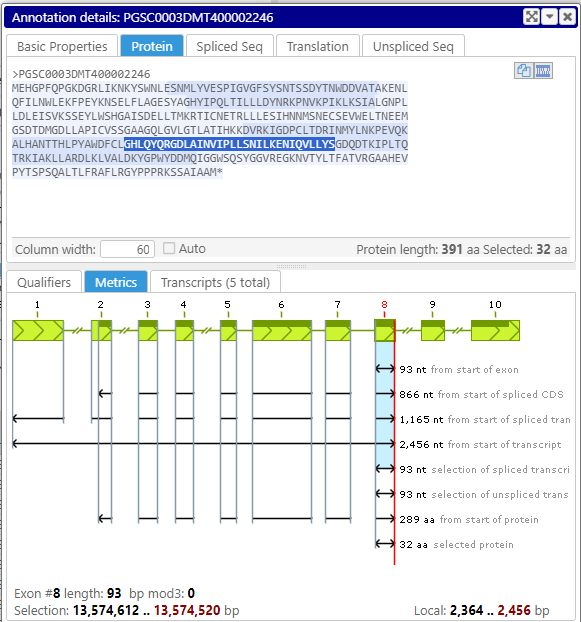
Protein tab 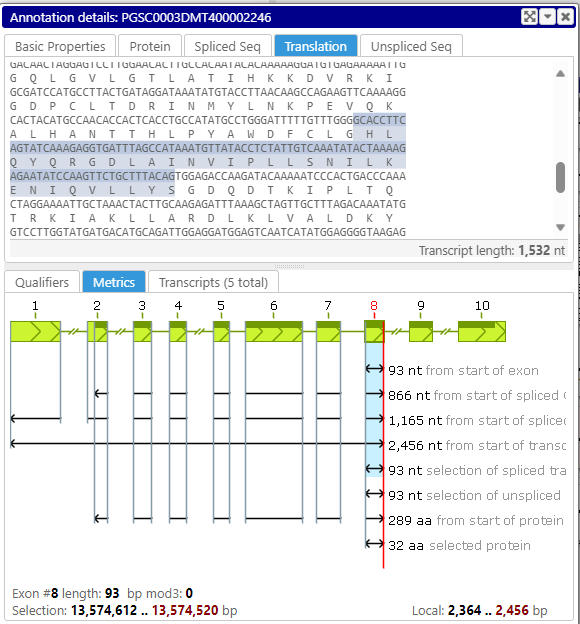
Translation tab
Save screenshots into SVG
We introduced a long-awaited feature frequently requested by the users – save screenshots as vector graphics suitable for publications. The SVG files will be created if you select an item under the Tools menu: Save SVG screenshot.
The vector graphics saves the image in the form of commands that draw primitives, such as lines or triangles. This means that you can view the images at any resolution, just zoom in when viewing the SVG file and the primitives will be redrawn using the saved instructions at a new scale.
Thank you for your valuable suggestions!



