DIAMOND as a much faster alternative to BLASTP
19:48 27 October 2021
in General News
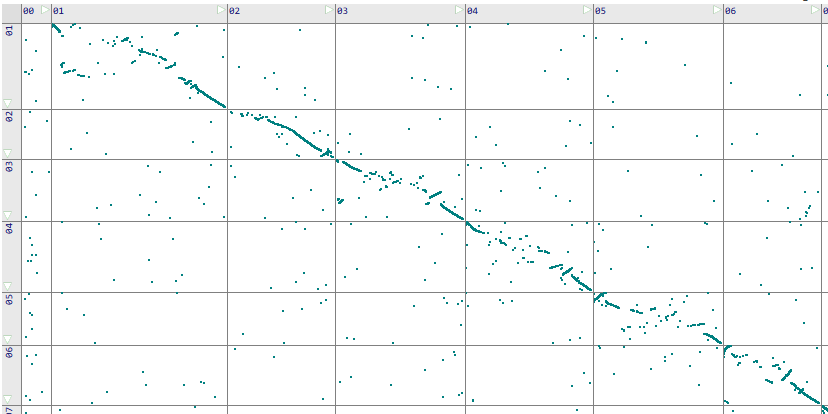
Our loader application PersephoneShell used to upload the data to the Persephone database provides commands to calculate ortholog gene pairs. By default, we use reciprocal BLASTP as the protein sequence aligner. A much faster alternative to BLASTP called DIAMOND (http://diamondsearch.org) can be chosen to speed-up the calculations. In our tests, by using DIAMOND to find the orthologs between two sets of ~30,000 proteins, we gain about 50x-100x performance improvement, retaining a similar sensitivity. For example, here is a comparison of synteny matrices for tomato and potato genomes produced by the two methods:

Reciprocal DIAMOND