26 Zea mays NAM lines in the pan-genome browser
We have added the data for 26 Zea mays reference genomes from the pan-genome project “Whole-Genome Assembly of the maize NAM founders” (https://nam-genomes.org/). The source files were downloaded from MaizeGDB.org.
To help browsing this pan-genome data set and provide the way to align the syntenic maps, we created artificial markers by cutting 200 bp sequence tags (spaced at 20,000 bp) from the main reference ZM-B73 genome. These “fake” markers were mapped onto the other corn lines, thus creating marker tracks on each map. Persephone automatically detects identical markers that appear on different maps shown on screen and draws a connector line. This approach allows linking the maps without engaging the massive ortholog pair records.

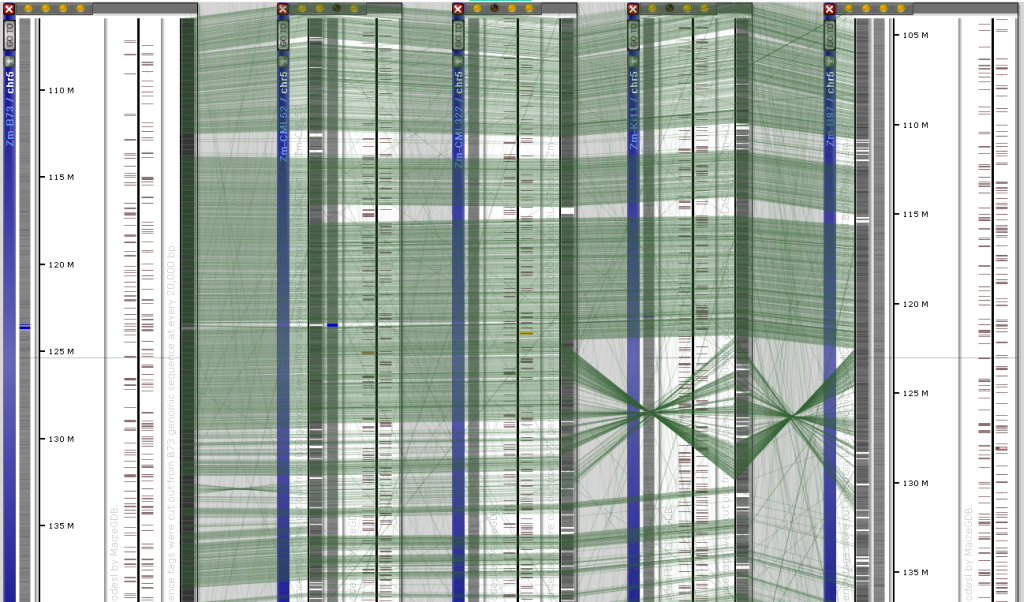
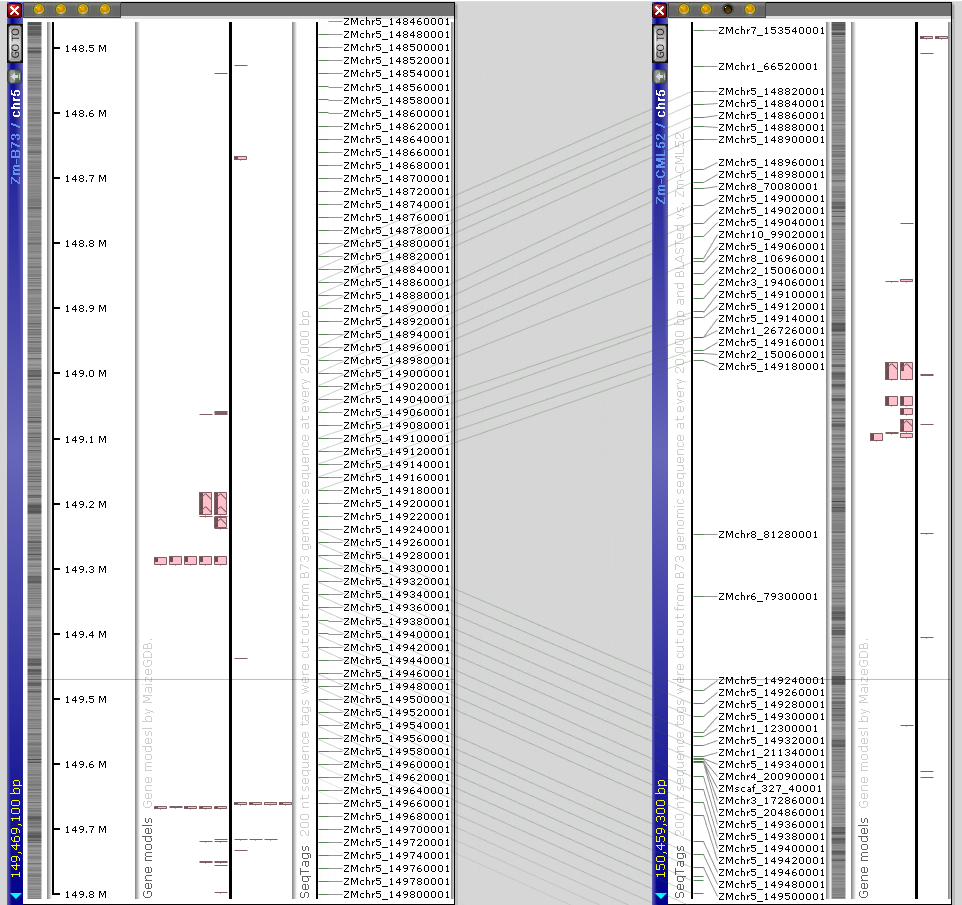
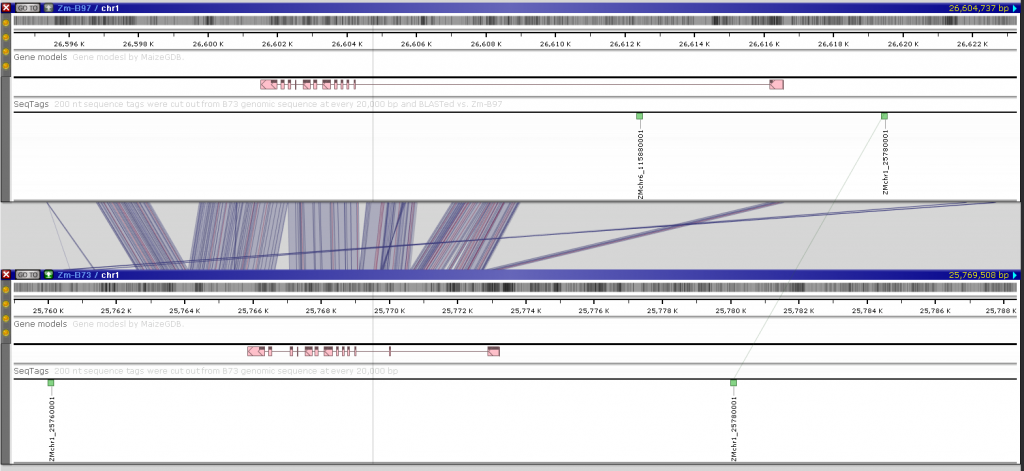
The Web version provides a new feature described previously, that allows a real-time comparison of the genomic sequences with one base resolution. You can use the connector lines to locate regions of possible sequence similarity, zoom in to view the sequence of 1 Mbp or less and press the “Instant BLAST” button to run BLASTN between the neighboring sequences. The fine-grain structural variations will be easily detected:
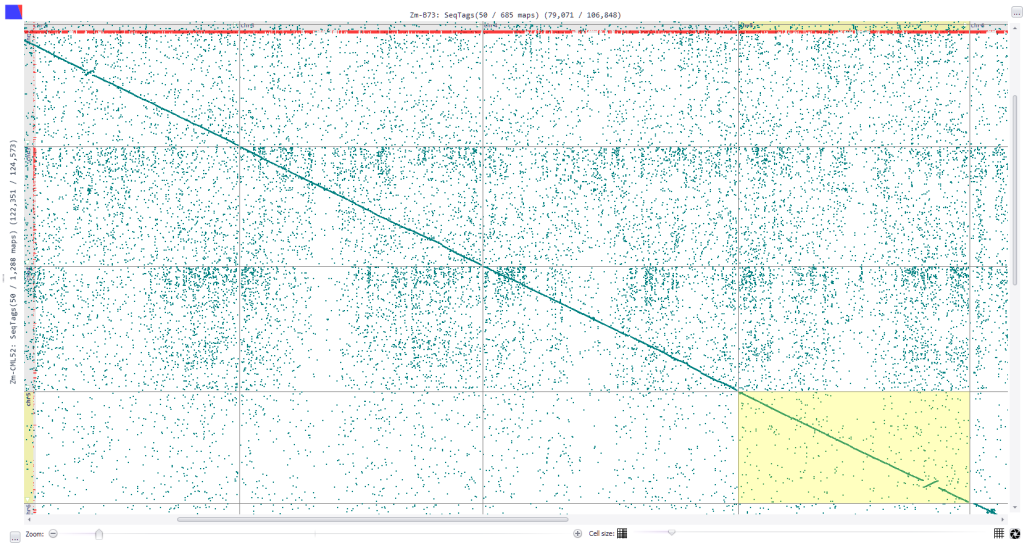
In addition to the tools above, the synteny matrix available in the Windows desktop version will help you visualize the entire genomes and the large scale rearrangements, which is especially valuable in the pan-genome context.