21 Reference Assemblies Added To The Rice Pan-Genome Collection
In collaboration with our colleagues from KAUST, we have added 21 reference-quality rice genome assemblies to our web portal and to the demo Windows version.
Os GJ-temp: IRGSP-1.0 (Nipponbare)
Os GJ-subtrp: CHAO MEO
Os GJ-trop1: Azucena
Os GJ-trop2: KETAN NANGKA
Os cB: ARC 10497
Os XI-1B1: IR 64
Os XI-1B2: PR 106
Os XI-1A: Zhenshan97
Os XI-3B2: LIU XU
Os XI-3A: LIMA
Os XI-adm: Minghui63
Os XI-2A: GOBOL SAIL
Os XI-3B1: KHAO YAI GUANG
Os XI-2B: LARHA MUGAD
Os cA2: NATEL BORO
Os cA1: N22
O. nivara
O. rufipogon
O. glaberrima
O. barthii
O. punctata
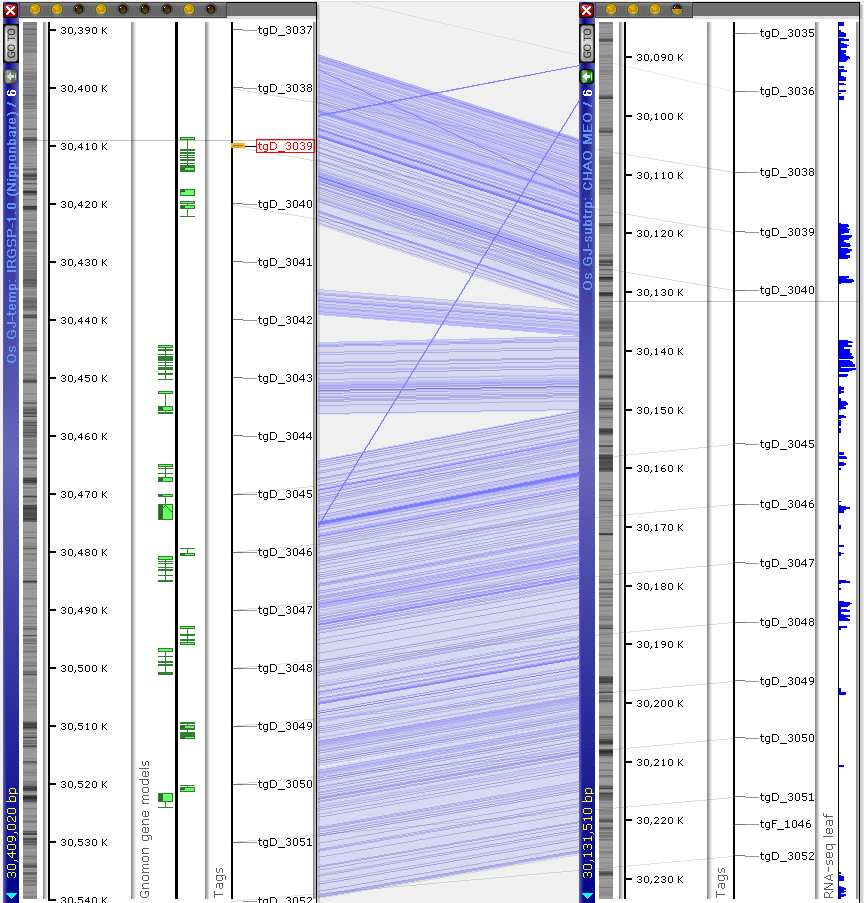
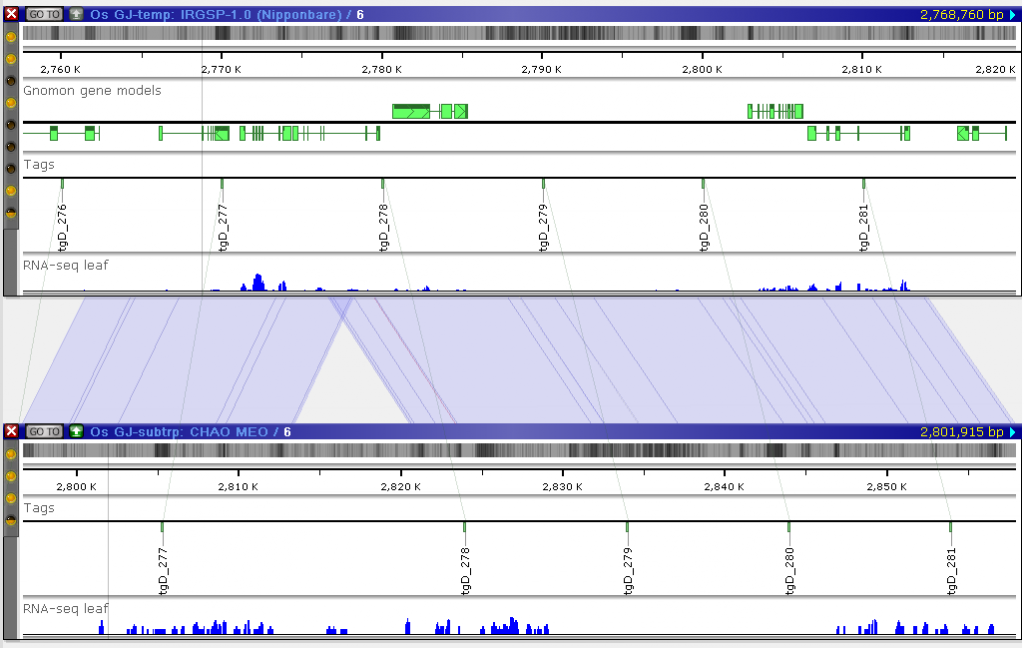
It is an ongoing project. So far, the sequences have been published without much annotation. Majority of the sequences have a few RNA-seq coverage tracks. As for the gene annotation, only the official Oryza sativa assembly IRGSP-1.0 has several tracks with gene models predicted using different methods. Meanwhile, the assemblies can be aligned by linking common markers. These markers have been generated by cutting out short sequence tags from the main reference IRGSP-1.0. Their locations in the other genomes were found by mapping them using BLASTN.
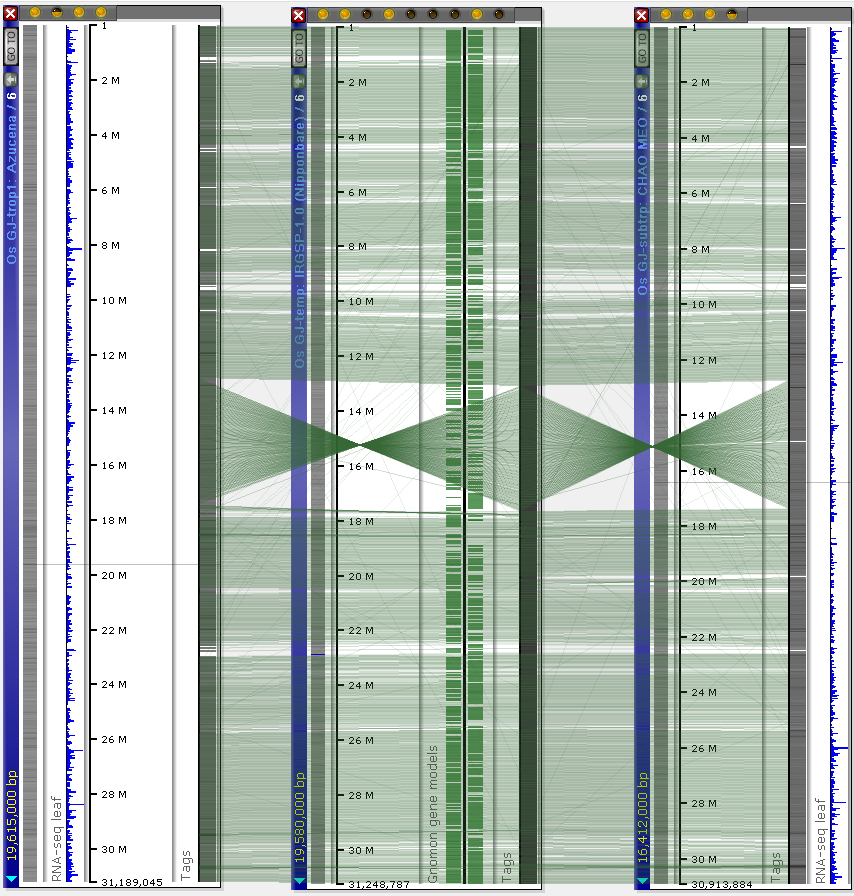
This alignment helps reveal the structural variations, such as the large inversion in the chromosome 6 of IRGSP-1.0. The small rearrangements can be further studied on the nucleotide level by zooming into the region of interest, using the connector lines as guides, and running the “instant BLAST” analysis on the visible part of the aligned sequences: