Web Version: Major Upgrade Introduces User Tracks
We rolled out a new version of Web Persephone with major feature upgrade: the users can upload their own tracks, and we made sure that all possible optimizations necessary to achieve maximum performance are in place. The system should handle files with millions of positions.
For now, we support user tracks created from marker files (csv, Excel, tsv) and – drum-roll – BAM files (see a separate post).
The files with marker mappings can contain various types of features. What is common for them – the features will be shown as named regions or points with genomic coordinates. These can be a list of locations of mapped markers, masked repeat regions, or other types of sequence annotation.
Once uploaded, the files will reside on our server and will be accessible from different computers.
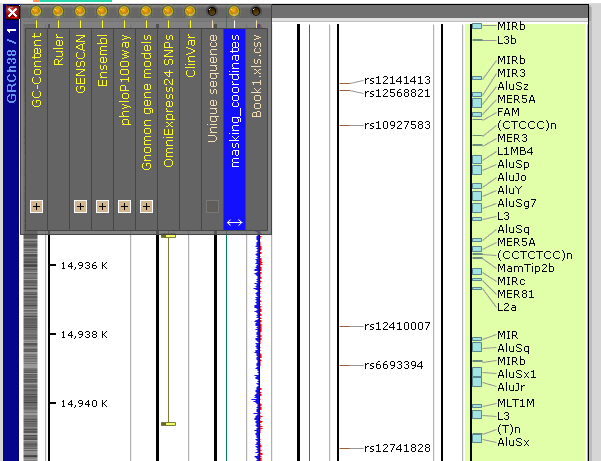
Here is how the track (highlighted) with masked repeats for human genome looks like (20,000 repeats on one chromosome):
The file format for marker data file is simple. We require 3 columns to be provided on each line with map name, feature name, and start (and, optionally, end) position: 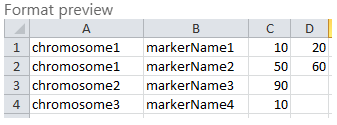
To be able to upload the tracks, the user must register by providing email and the password. Each user will get a free disk space of 200 MB on our server.
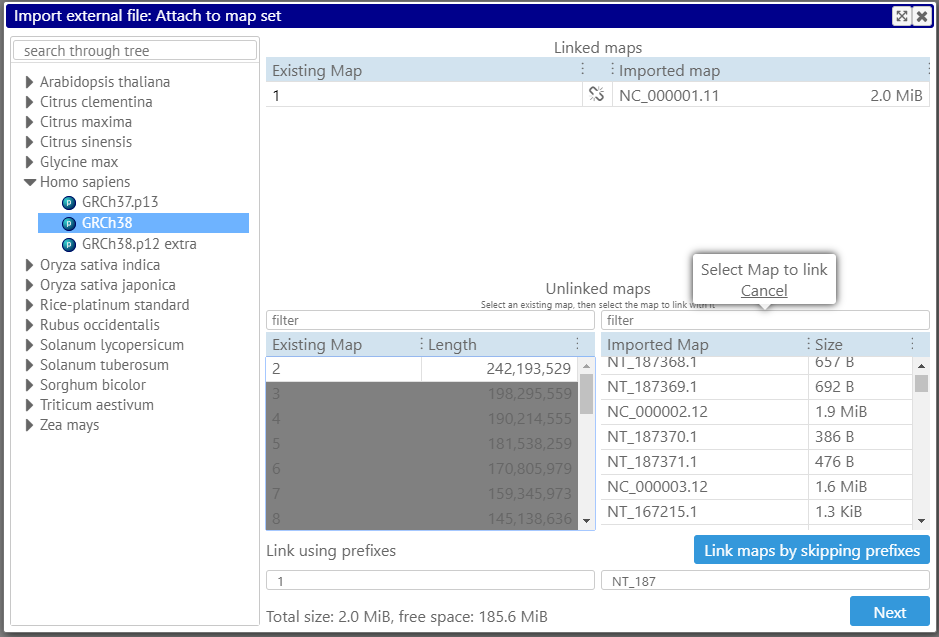
The procedure of loading the files, besides selecting the file type, includes matching the map names from the file to the ones in the database. If the maps use the same names as in the database, there is nothing to worry – Persephone will display the data on the corresponding maps. However, if the maps use a different naming pattern, e.g., “chr.1” instead of “Chr1”, the program will try to match the maps automatically and will provide a way to correct the name correspondence manually. In cases when the matching is not trivial, linking the map pairs still can be done, but only manually. For example, if you want to match the map called “2” to “NC_000002.12”, select these maps in the lists on the left and on the right:
Please see a separate post about visualizing BAM files.