Transcript structure comparison view added to the Web version
The gene properties form shown for a selected gene model has now a new tab, called Transcripts:
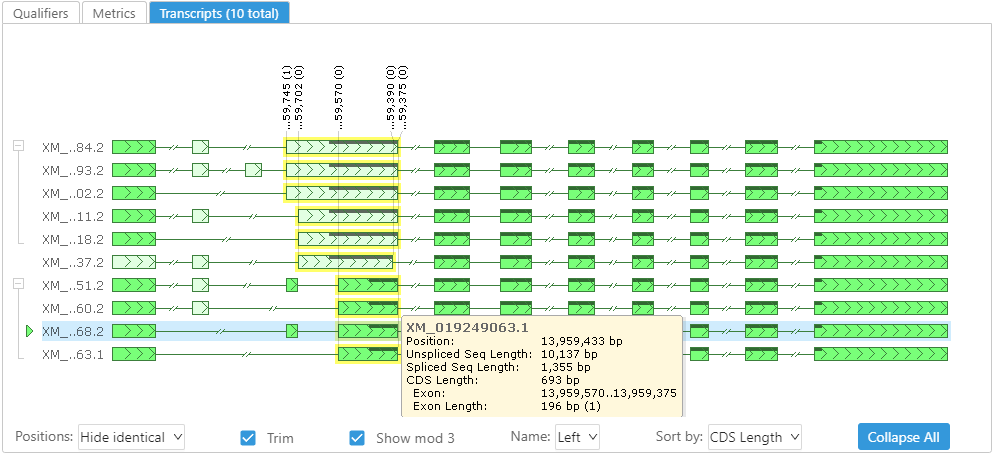
This control displays alternative splice variants with corresponding measurements. A reference isoform (highlighted) is compared to the other variants. Whenever an exon is different from the reference, it is painted with a lighter color. This makes it easier to spot the differences in the gene structure. It is possible to hide all the measurements or leave only those that are different. If you move the mouse over an exon or an intron, the corresponding numbers will pop-up. We find it useful also showing the remainder of division of the sizes and coordinates by 3 – the size of a triplet – so that it is clear if the missing exon shifts the downstream reading frame, etc.
The transcripts can be ordered by name, unspliced sequence length or the CDS length. The models sharing common CDS can be grouped and collapsed to display all distinct coding reading frames.
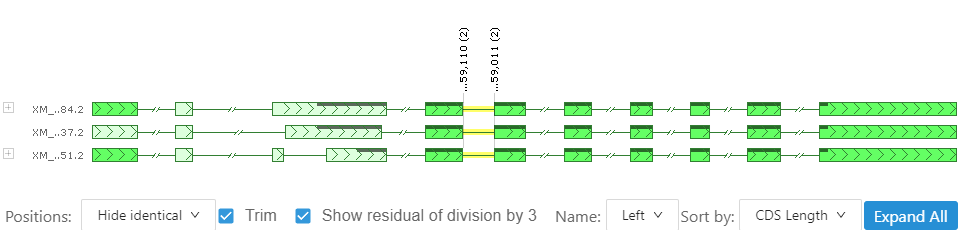
This interface allows panning and zooming, so you can see more details and focus on an area of interest.



