The Spring Update: Drag&Drop VCF and Multiple Sequence Alignment
The Persephone version, published recently, presents a few improvements.
Drag-and-drop VCF files.
Users are now able to import their VCF files. The remote files are referenced by a URL, and the local files can be added via drag-and-drop. For files larger than 15 MB, we require an index file (with extension .tbi or .csi). Read more…
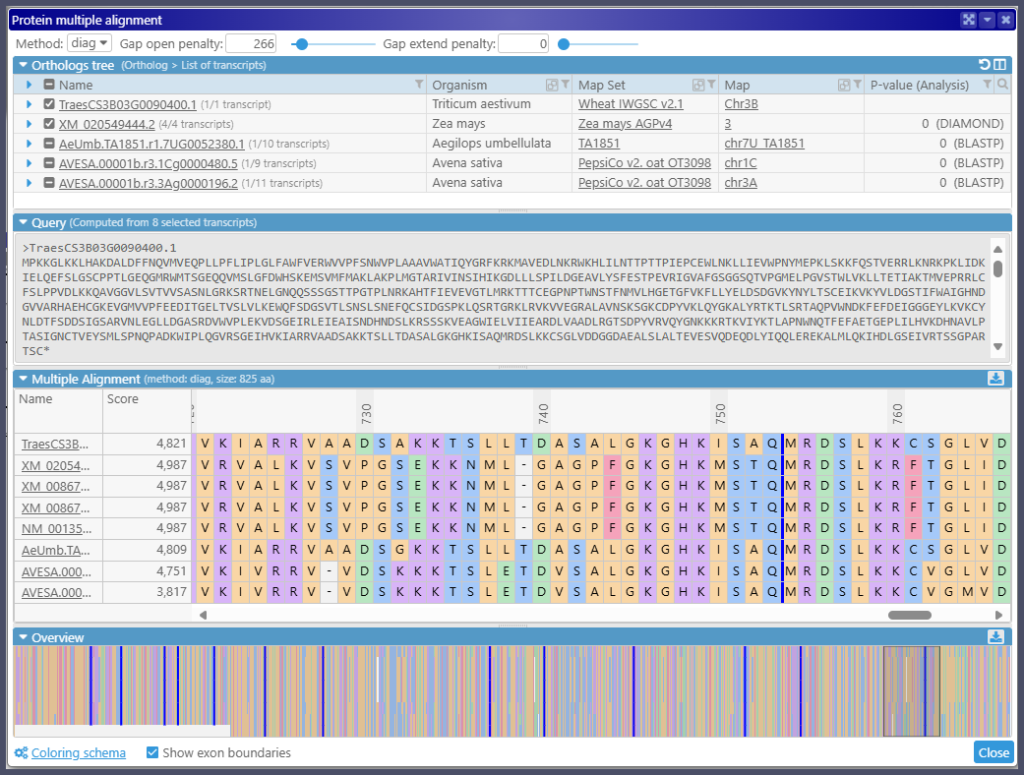
Multiple Protein Sequence Alignment
The new tool aligns multiple protein sequences. You can access the alignment interface via the Tools menu. In this case, the text box for the sequences will be empty, and you can populate it with the sequences you wish to align, most probably from the clipboard. Alternatively, you can open the alignment tool via the gene properties form. The Orthologs tab now has a hyperlink to show the alignment interface pre-populated with the orthologous protein sequences. Read more…
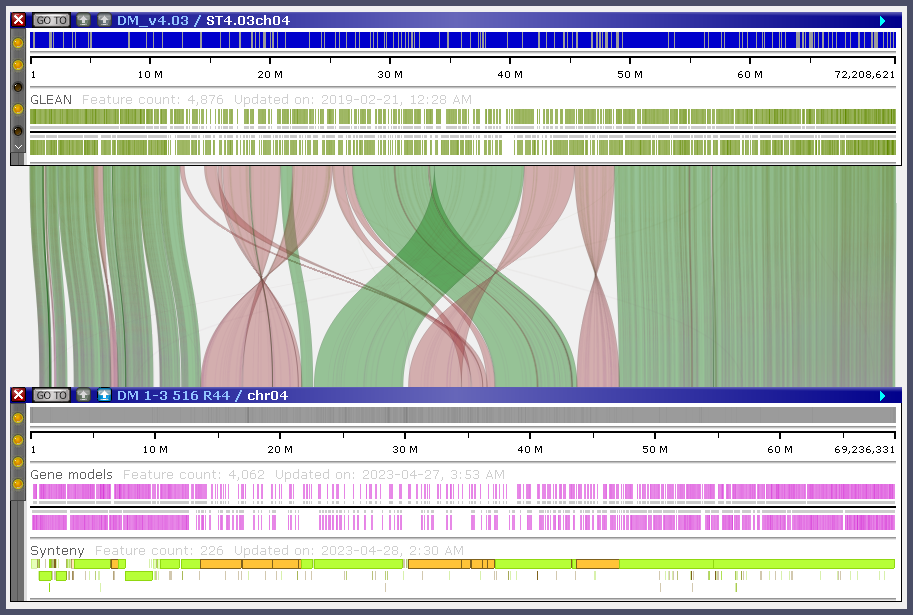
Curvy connectors
We decided to spend time on the curvy lines and ribbon connectors as we believe that scientists will appreciate the ability to produce publication-quality graphics without engaging external tools. The curves not only enhance visual elegance but also offer a more informative representation.
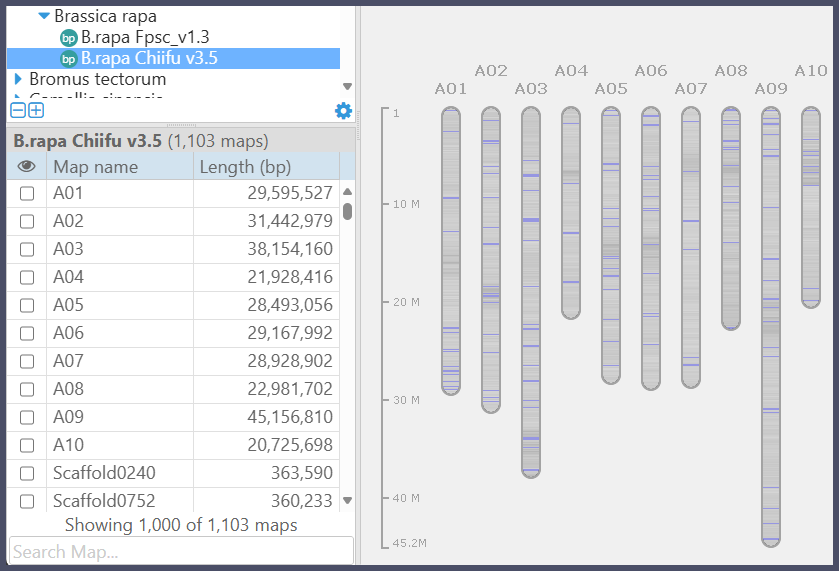
Sequence histograms in the Bird’s-eye view of genomes
As you probably noticed, we show a graphical representation of the selected genome when the Persephone’s stage is empty. We made the overview of the maps more informative by adding the sequence histograms. They are based on the GC content of the sequences and indicate the locations of the poly-N spacers, shown in blue.
Importing and indexing GFF files
When users’ GFF files are imported via drag-and-drop, the features are indexed for search by default. You can now skip this step if you are not planning to run BLASTP or search for text keywords, which will save you some time.
When matching the maps in the file to the maps in the database, you can now establish the link using the map’s accession number in addition to the map name. Read more…
Detect variants that disrupt splicing
Our Translation Changes interface highlights variants that lead to modified protein sequences. In the latest update, Persephone also flags alleles that affect splicing sites or disrupt start and stop codons, providing deeper insights into translation changes. Read more…
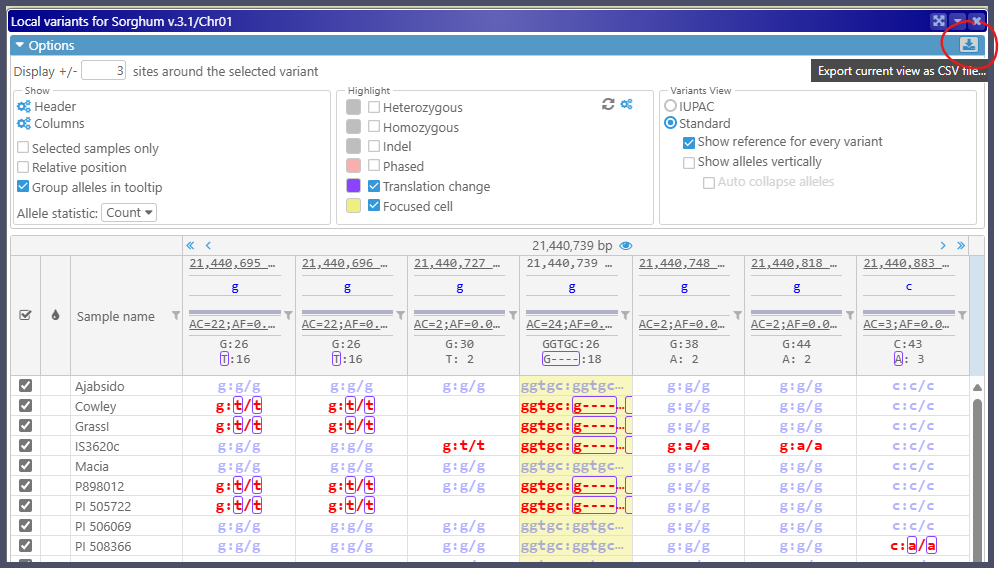
Export the Local Variants table
The Local Variants form shows the neighborhood around the clicked variant, including all available samples and adjacent variant sites. Each cell can be formatted in many different ways. The new feature allows users to export this table in the currently formatted view to a CSV file, which can be opened in Excel.
Please try the new features and let us know what you think.
Thank you.



