The New Year Update: improved pangenome visualization.
Happy New Year, All!
We are going to the Plant and Animal Genome conference in San Diego. There, we will present our latest version of Persephone. It includes new features that make navigating pangenomes easier.
We are introducing the redesigned Synteny matrix, Multimap, docking windows, and gap statistics for BAM reads.
A frequent question for those who work with pangenomes is how to find the best map to match the selected one. This can be challenging if your data set contains hundreds of millions of unassembled scaffolds.
Assume you have a hundred genome assemblies for closely related species. If the sequence similarity is good, you can link the corresponding maps by common markers. Persephone can generate short sequence tags at the back end by cutting out subsequences and mapping them onto other assemblies. The link can also be established via orthologous gene pairs. Now you might want to ask a question: which map has a similar set of features that can be linked to my map?
In Persephone, finding the matching map can be done in a few ways.
“Find synteny” menu item
If you already have the map of interest on the screen, right-click on a track with markers or gene models and select the menu item “Find synteny…”. This will open the Multimap interface:
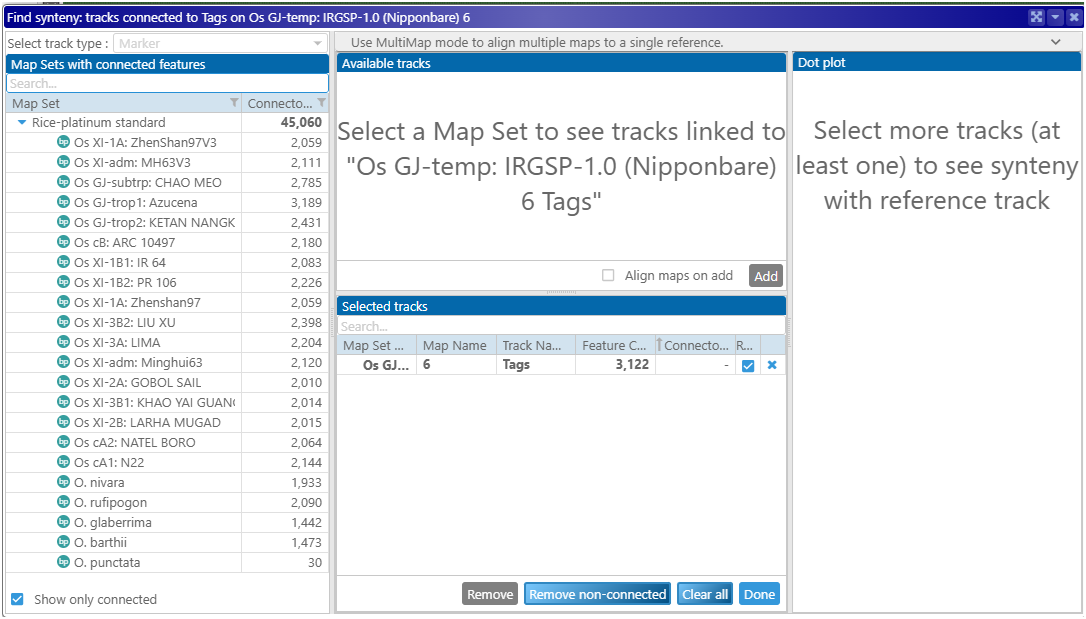
The clicked track becomes the reference (the panel “Selected tracks”). The tree on the left shows genomes that could be linked to the reference by common markers (we started with the marker track “Tags”). The genomes that cannot be linked can be hidden.
The connector counts column will help pick the proper genome. Let it be CHAO MEO:
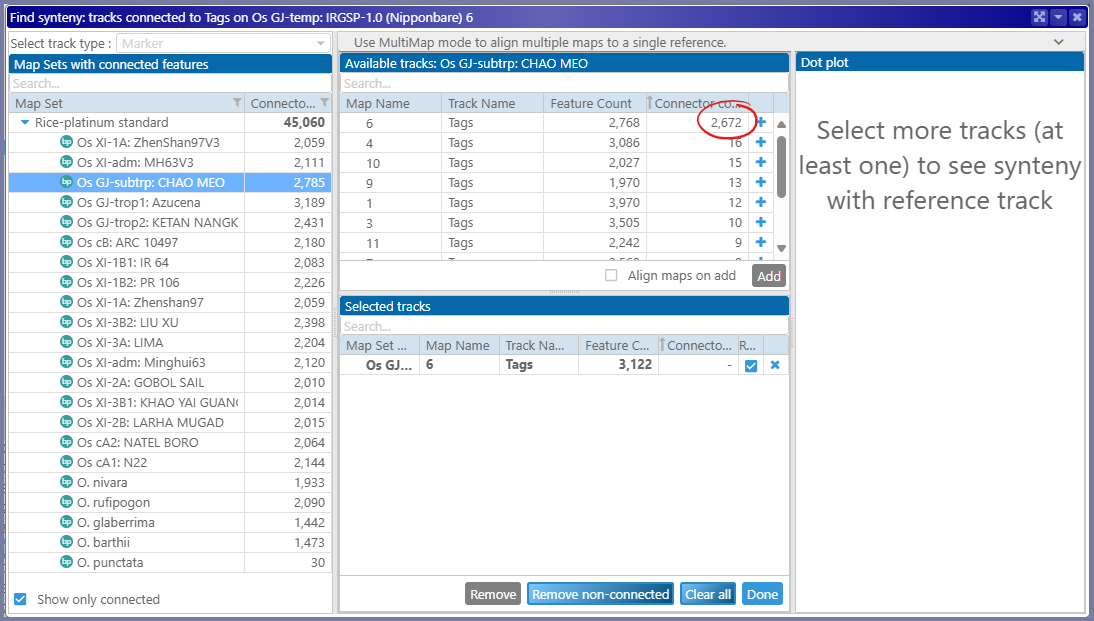
The best matching map will appear at the top of the list of available maps/tracks. Adding it to the list of selected tracks will add the corresponding map to the stage. The other tracks will remain hidden, but you can always add them manually.
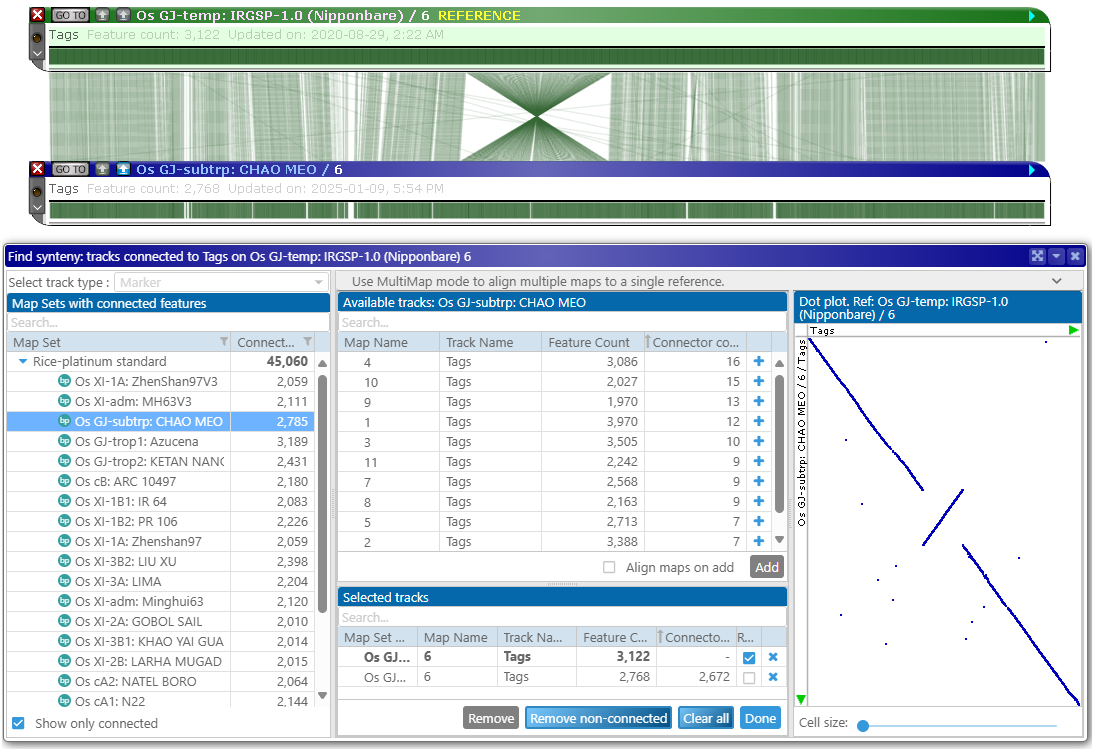
You can continue adding more maps. If a reference map represents an assembled chromosome, the matching scaffolds from other assemblies will automatically be placed at the corresponding location:
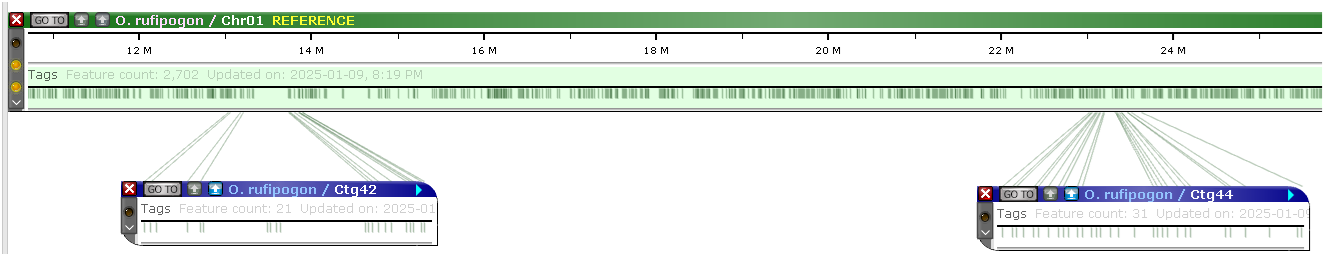
Multimap
The Multimap interface is designed to select matching maps quickly, even if there are millions of candidates. By leaving only relevant tracks and hiding the others, the Multimap mode allows users to stack many maps on the screen. The interface controls will highlight the presence/absence of features and bundle the individual connectors into ribbons to simplify the picture. The locations of individual features can be traced:
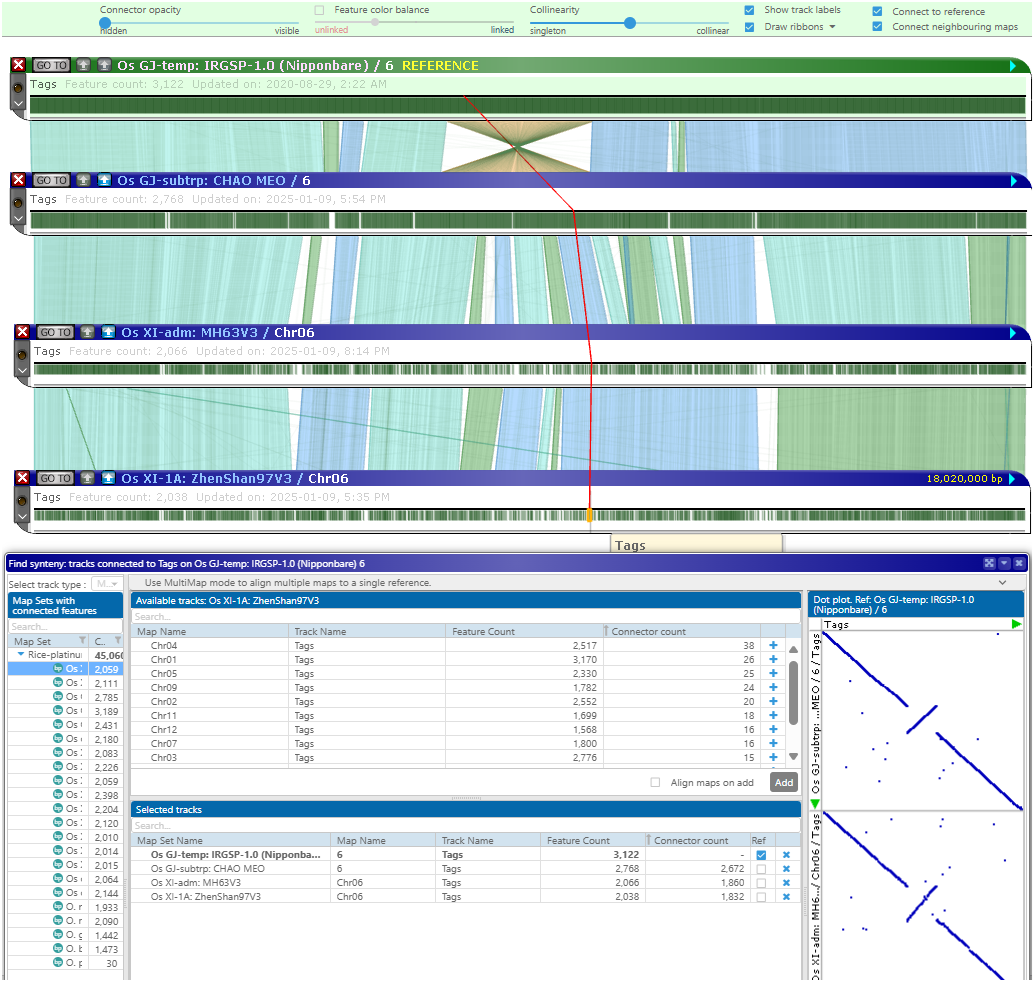
A special design of the data structure generates the counters of common features and finds the matching maps in real time.
Redesigned Synteny matrix
We have also redesigned the Synteny matrix. It now shows the number of connected features for all maps in the genome combined. While the Multimap interface finds the matching tracks for a selected track on a single map, the Synteny matrix compares entire genomes based on data in the selected track names. In addition to the gene models and markers, the track types shown in the matrix now include the synteny ribbons that link sequences with high similarity.

After the track has been selected in the top “query” panel (X-axis), the bottom panel (Y-axis) will show only the genomes/tracks that can be linked to it. This greatly facilitates navigation through multiple interconnected genomes.
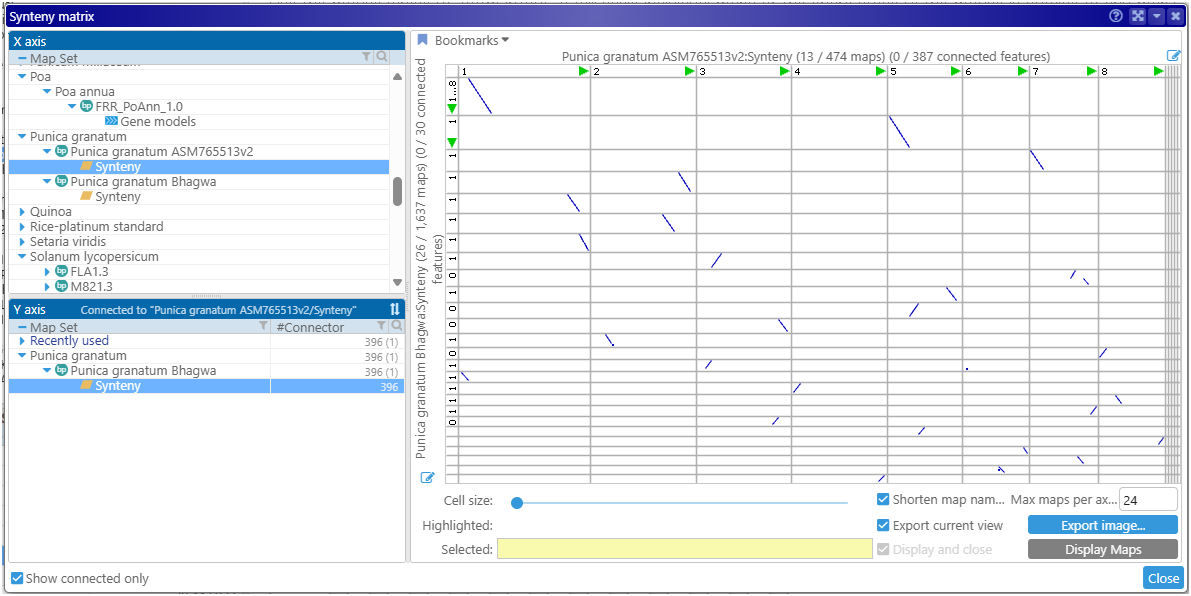
Gap/insertion statistics
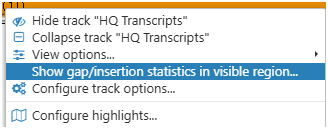
If you work with the NGS reads, you might be interested in analyzing certain sites targeted by CRISPR. We added an interface showing how many mutations of specific sizes are captured in the reads mapped to a region. You can see how many reads confirm deletions or insertions of different sizes. Call this function by right-clicking in a bam/cram track:
Docked windows
The main interface has been upgraded: forms can now be docked to the edge of the screen and reused. For example, you can view marker details for different markers in the same window. Once docked, the form will remain fixed on the screen’s edge, updating its contents with each marker click. The form does not have to be docked to be reusable. A separate “pin” control allows it to be reused in a “singleton” mode:
Tiny font
We added one more size of the interface font: “Tiny”. If you feel that the forms or grids appear too crowded, try pressing the shortcut Alt-1 to switch to the Tiny font. The other sizes can be selected by pressing Alt-2, Alt-3, Alt-4, or by choosing the font from the drop down menu in the Settings/Display.
Please visit us at the booth #222 in the exhibit hall. We will run the Persephone workshop on Tuesday, January 14th, at 1:30 pm, Palm 1-2.
Thank you!



