The New 2026 Year Update: Structural Variants, Multiple Alignment of BLASTN Results, etc.
The Persephone version, published recently, presents a few improvements.
Handling BCF Files
In addition to VCF, Persephone now directly supports BCF files imported into the main application. Users can add variants by referencing remote BCF files (with an index) or by loading local files.
New Functionality for BAM Tracks
We significantly increased the limits for the data displayed in BAM/CRAM tracks. There can be genomic regions with many mapped reads. Persephone can now handle peaks of up to 300,000 reads. Naturally, not all will fit the screen, but statistics for each site will be calculated from an analysis of all available reads.
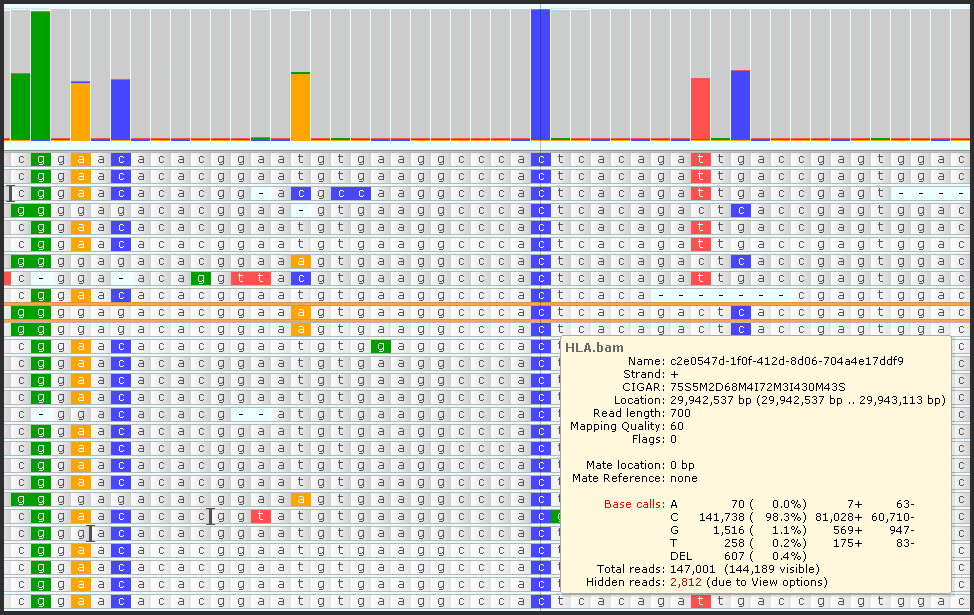
To facilitate handling this amount of data, we introduced down-sampling, filtering, and sorting the reads. Read more… Please also check this.
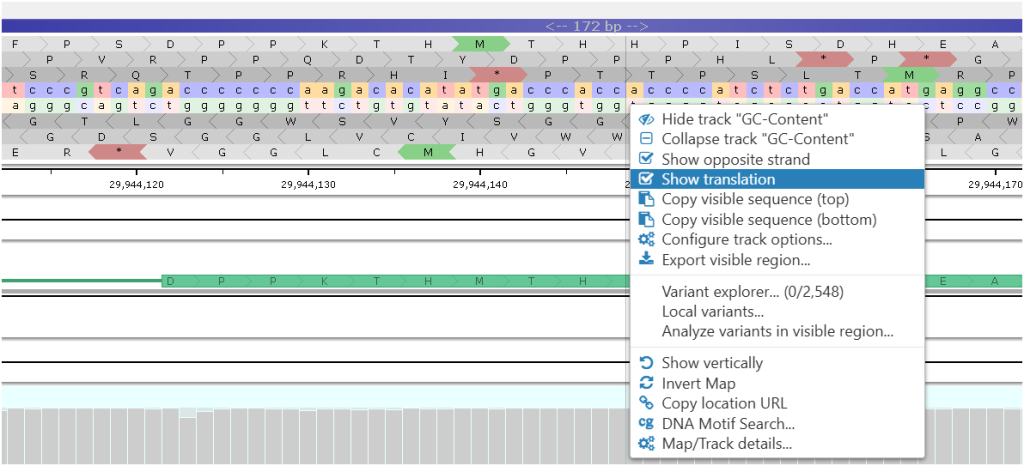
The Sequence Track Can Show 6-Frame Translation
At a certain zoom level, when individual bases are visible, we provide an option to display the genomic sequence translation in 3 frames. Showing the opposite strand can also add three more translation frames. The start and stop codons are marked accordingly.
Fine Control Over Data Displayed in Tables
Persephone uses tables in multiple parts of the interface. We have added extra context menus to most of them, providing control over the way the data is displayed. For example, users can now edit columns by selecting checkboxes in the list of available columns, export individual columns or rows, etc. Read more…
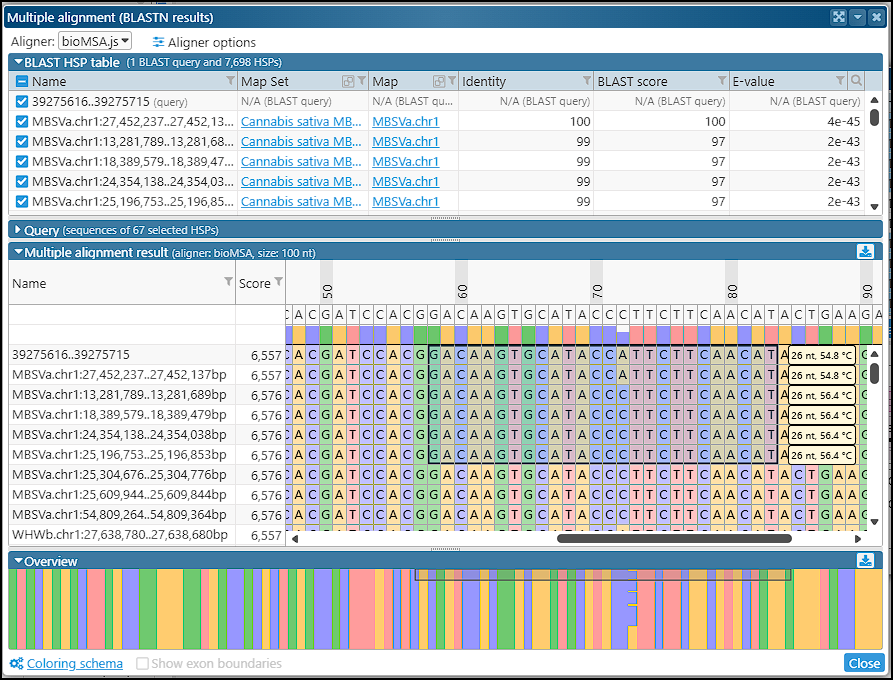
Multiple alignment for BLASTN results
We have added a helpful enhancement to the BLASTN results form: the subject sequences for each HSP can be included in the multiple alignment interface. For instance, you can select a region of interest and use its sequence as a BLASTN query to find similar sequences in other closely related genomes. Read more…
Dark Mode Introduced
You might like the latest enhancements of the Persephone UI. The users have been able to adjust track feature colors and change the font size. In the new version, we introduced the Dark Mode. Read more…
Multiple Motif Search and Restriction Sites
We have improved the motif search interface, enabling users to search for multiple motifs by entering them as a comma-separated list. Additionally, users can now select DNA motifs from the REBASE database, which contains recognition sites for restriction enzymes. Read more…

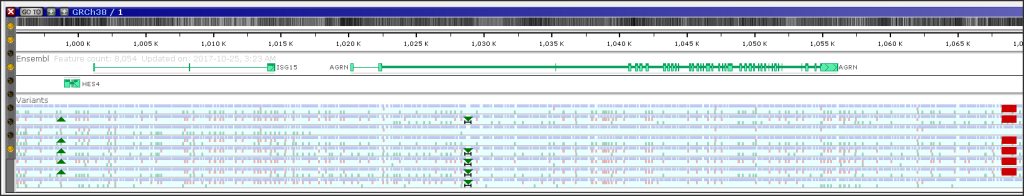
Visualization of Structural Variation
VCF files used to load variation data may contain symbolic allele notations, such as <DEL>, <INS>, or <DUP>. These symbolic alleles appear in the ALT field of a VCF record and are used when the actual inserted or deleted sequence is too long or not explicitly included.
The new version of Persephone will show such structural variants (SV) in the Variants track:
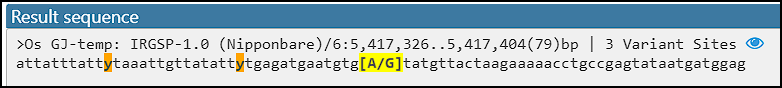
Design SNP Assay
If you design an assay targeting a specific allele, it is important to know the genomic sequence neighborhood. A new addition to the Local variants interface will allow users to export flanking sequences around the selected variant. The extracted sequence can be formatted differently and typically will use IUPAC codes for variant sites in the flanking sequence while splitting the alleles of the ‘anchor’:
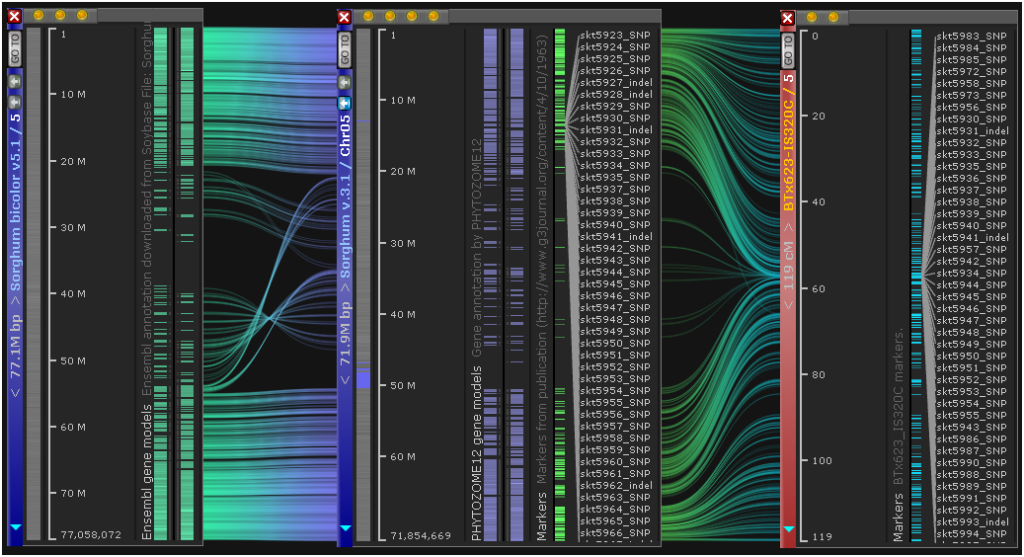
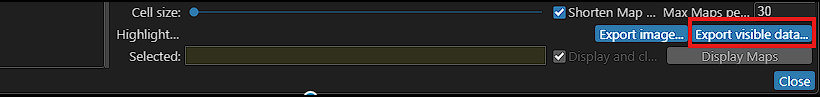
Export Ortholog Records
Synteny regions between maps are revealed by linking orthologous genes or common markers. Now you can export these pairs by clicking the button Export visible data in Synteny matrix form. Select two genomes, optionally zoom in on a region of interest, and export the orthologs or marker pairs with their selected properties.
New Genomes
We continue to add new genomes to our system at https://web.persephonesoft.com. Recently, we have loaded pangenomes for Cannabis (97 annotated assemblies) and Solanum (23). You can now find three dog genomes, finger millet, mouse, gorilla, celery, soybean Wm82.a6.v1, and others. The full list is available here.
PAG33
If you happen to attend the Plant and Animal Genome in San Diego, please visit our booth (#122) and the workshop (Tuesday, January 13th, 1:30 PM, Palms 1-2).
We will be happy to share the latest information and hear your thoughts.



