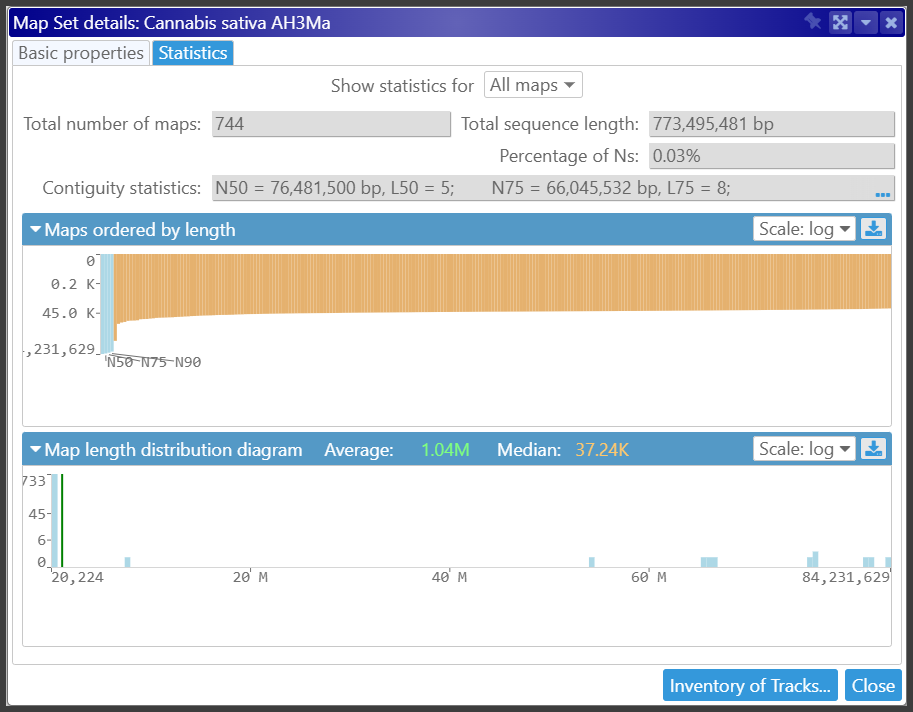
The Cannabis Pangenome Is Available
The Cannabis pangenome from the Michael Lab at the Salk Institute is now available in Persephone. With previously loaded genomes from NCBI and CSGDB, our collection has 97 assemblies with gene annotation.

We also calculated orthologous gene pairs, which allow us to align the maps from different accessions. This analysis resulted in a data set of 300 million orthologous gene pairs.
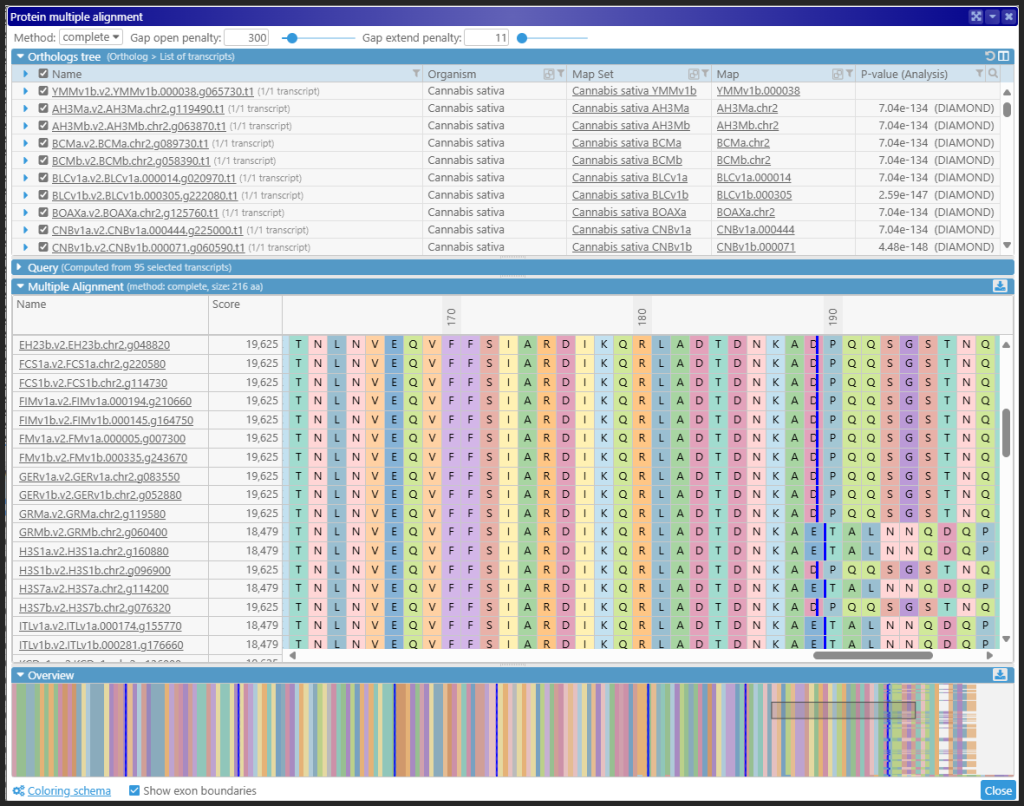
The protein sequences in the ortholog groups can be shown in our multiple sequence alignment interface. On average, each gene has 100 orthologs, and their structure is quite consistent.
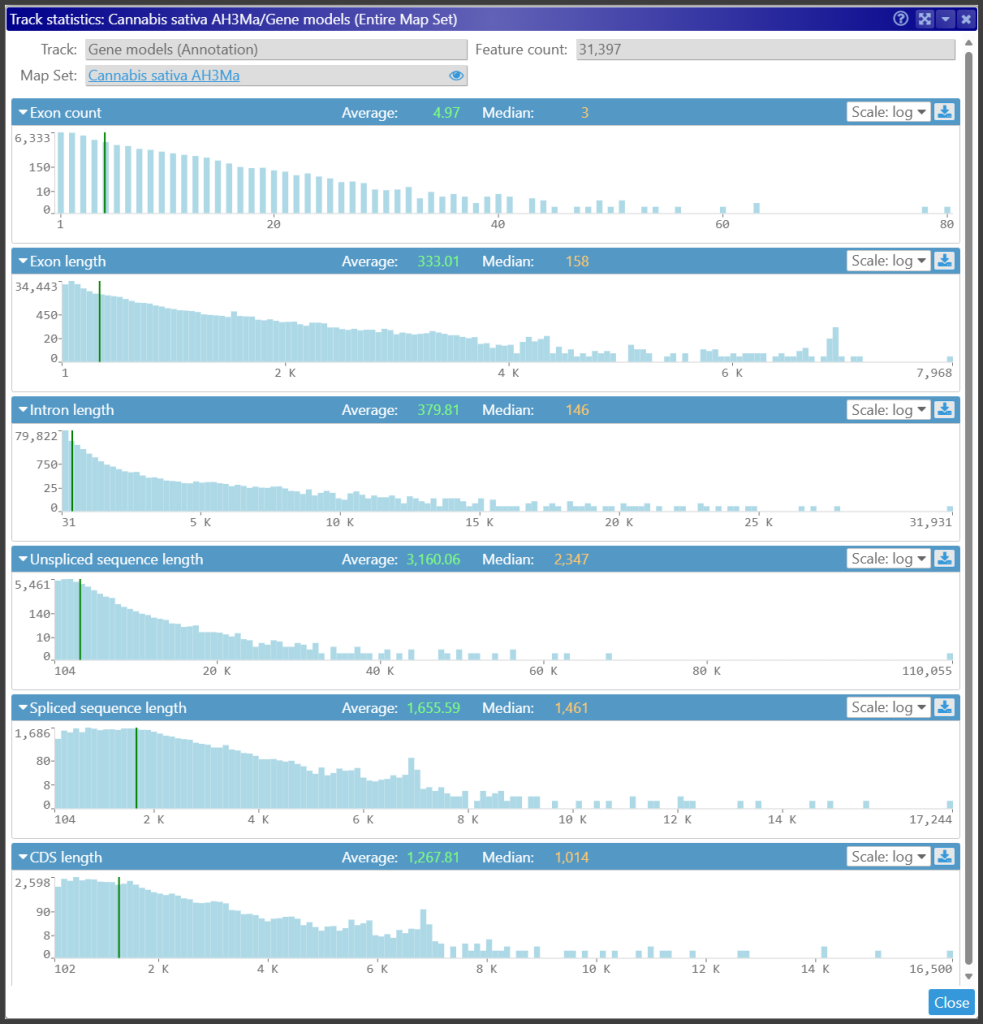
On a side note, we checked the predicted gene models and found them to be of high quality. Very few of the produced protein sequences have problems, such as the first amino acid not being Methionine. The exceptions appear mainly because the underlying sequences we loaded are masked, and the translation starts in a polyN region. Probably, the gene models were predicted on non-masked sequences. Here is an example that shows various distributions for gene structure elements. As with all genomes in Persephone, such a form with statistics is available via map set properties.
Also, we ran a quick assignment of gene function by finding the best match to SwissProt proteins.
By the way, the genomic sequences have very few polyN regions. Here is a typical summary of an assembly: