Solanum lycopersicum SL4.0 (tomato) genome browser
We have downloaded the assembly and annotation of the reference tomato genome version SL4.0, as well as other tomato varieties such as M82 or FLA.8924. The data is coming from http://solgenomics.net.
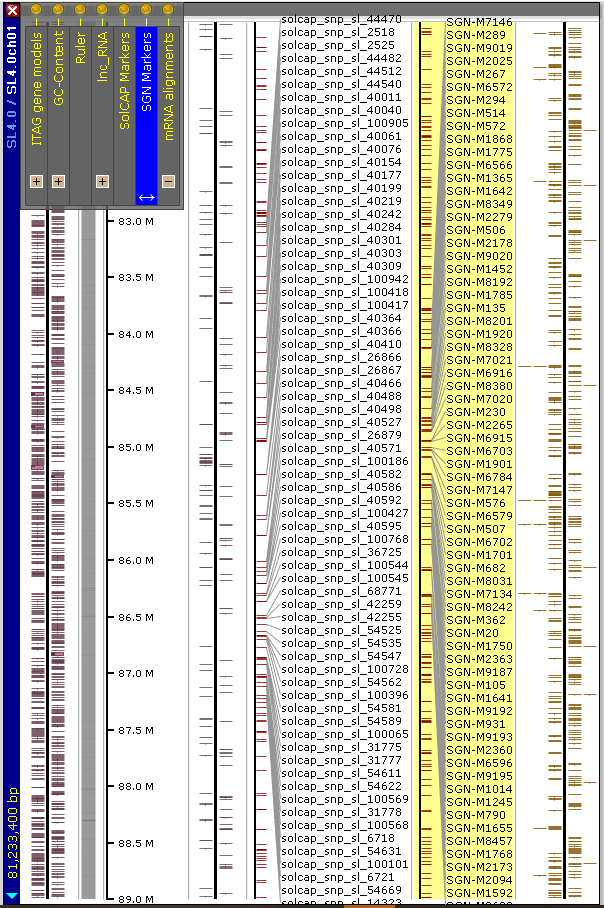
For now, the new map set has the following tracks:
- ITAG4.0 gene models predicted by maker
- Long non-coding RNA
- SGN mapped markers
- SolCAP markers
- spliced alignment of 300K tomato mRNAs from NCBI
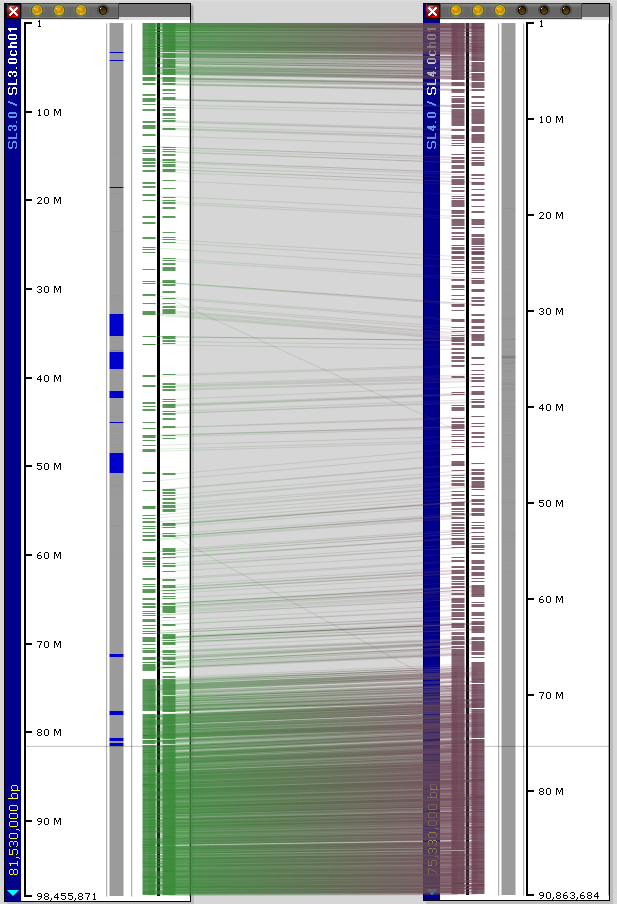
When SL3.0 and SL4.0 genome assemblies are aligned, Persephone helps visualize the changes between the versions. The blue regions in the picture below designate the poly-N spacers. Thin greenish hairlines link the gene models with highest similarity. For example, here is the comparison of the two versions for chromosome 1.
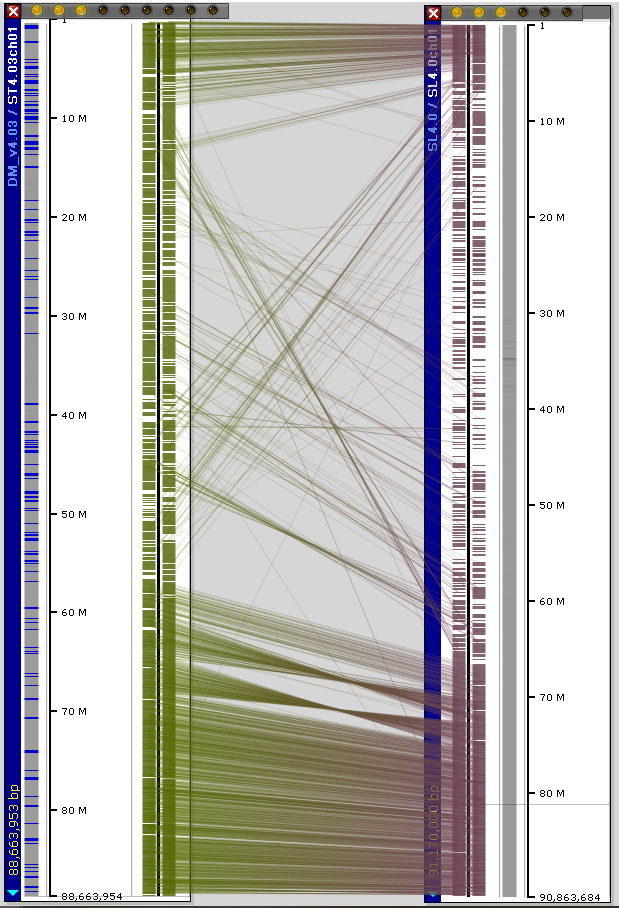
We also calculated the orthologous relations between the tomato (SL4.0) and potato (DM_v4.03) genes.
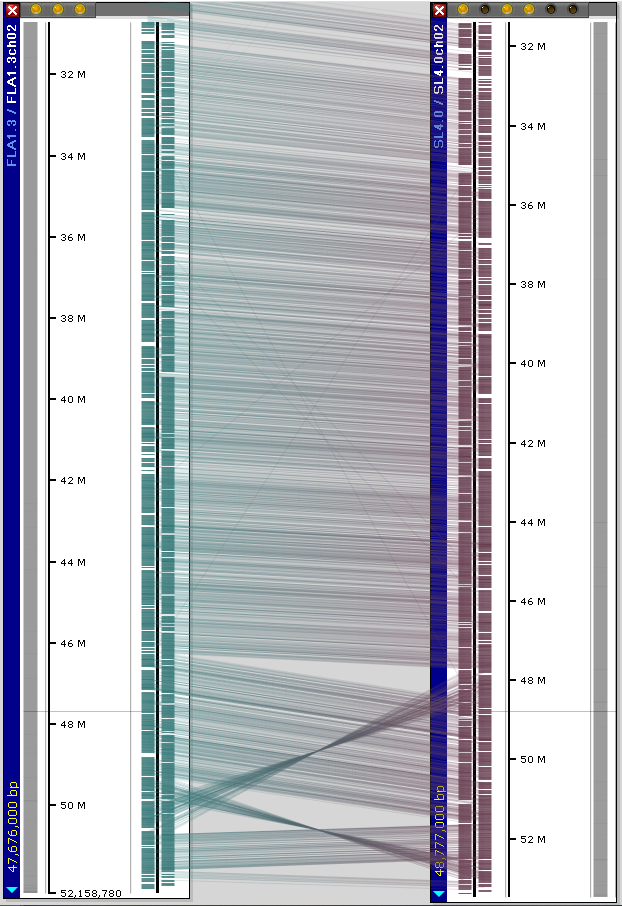
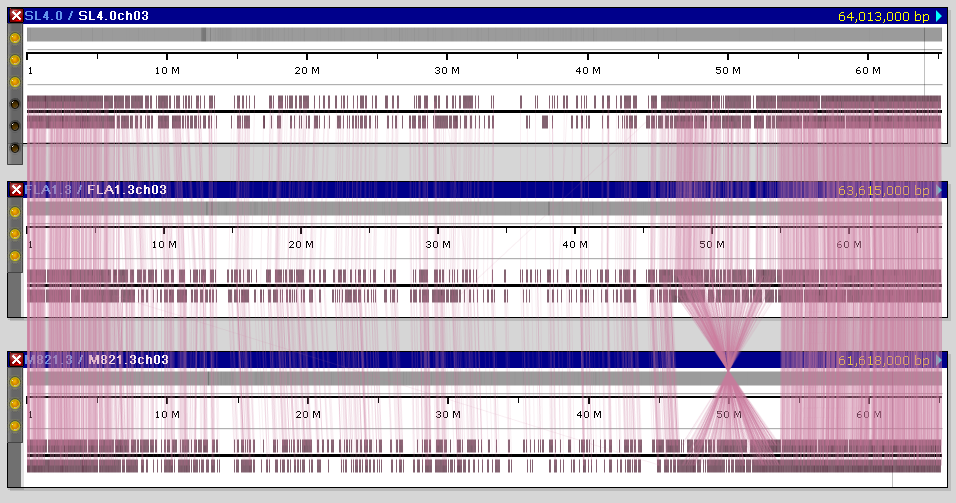
In addition to the popular reference genome SL4.0, we have loaded the assembly and annotation of varieties FLA.8924 (version FLA1.3) and M82 (version M821.3). The alignment of maps sometimes shows some structural rearrangements:
Please let us know if you would like to see more tracks on the tomato (or any other) genomes.