New data set: Wheat 2.8 M SNPs on IWGSC v2.1
We have added a new set of SNPs for the wheat genome (IWGSC v2.1). The VCF file with the variants (ExomeCapture) was downloaded from https://zenodo.org/records/7852690. According to the description of the dataset at Variations – WHEAT URGI (inra.fr), “VCF file of 2,799,166 single nucleotide polymorphism (SNP) markers positioned onto the Chinese Spring reference assembly RefSeq v2.1 developed by the International Wheat Genome Sequence Consortium (IWGSC; Zhu et al., 2021). These SNPs were lifted from the 1,000 wheat exome project, originally positioned onto RefSeq v1.0 (He et al., 2019). The SNP projection from RefSeq v1.0 onto RefSeq v2.1 was accomplished using LiftOff (Shumate and Salzberg, 2021).“
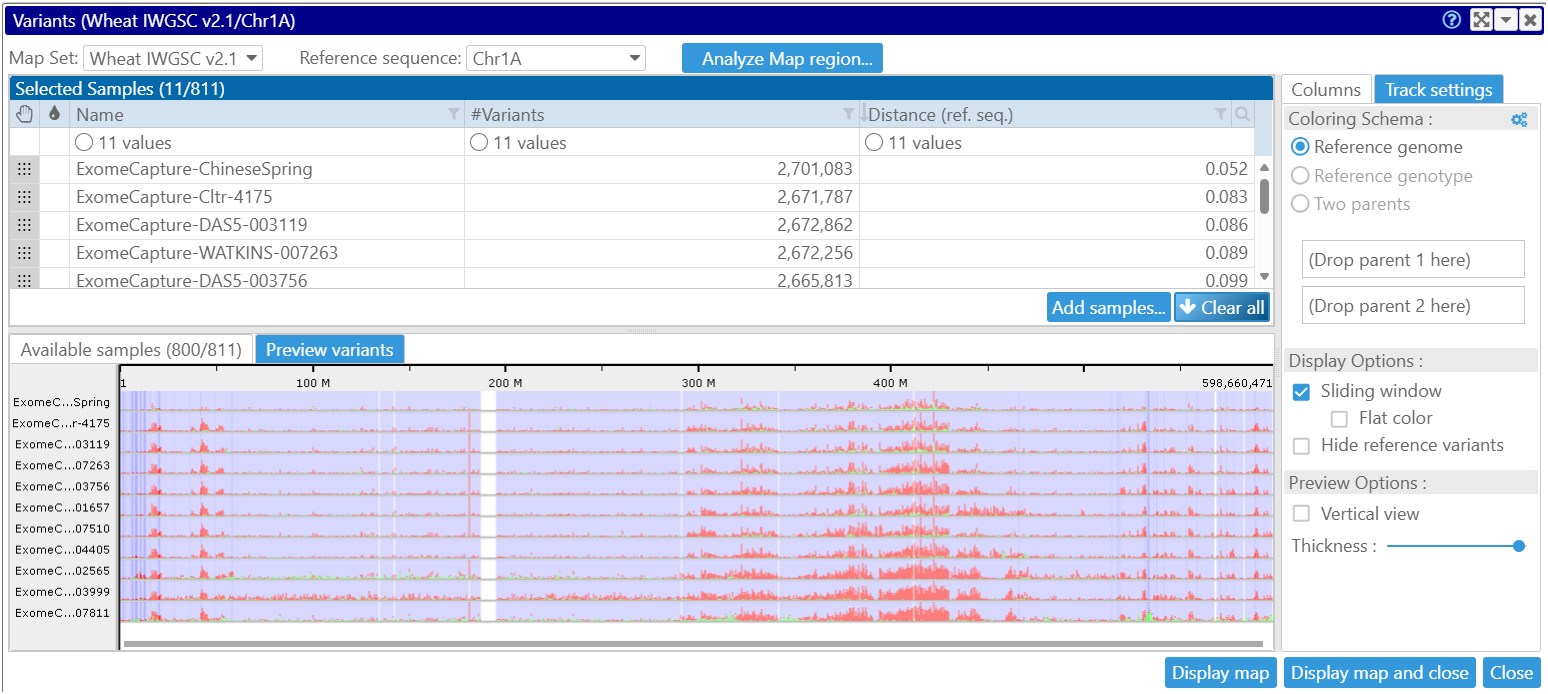
Our web portal stores 811 genotyping samples which can be visualized side by side. The sample selection can be done based on the genetic distance to the reference sequence or to a selected parent variety. Persephone will visualize the regions where the samples are significantly different. As usual, it allows sorting the samples and zooming in to see the individual alleles. A specialized interface will highlight the translational consequences of the mutations.