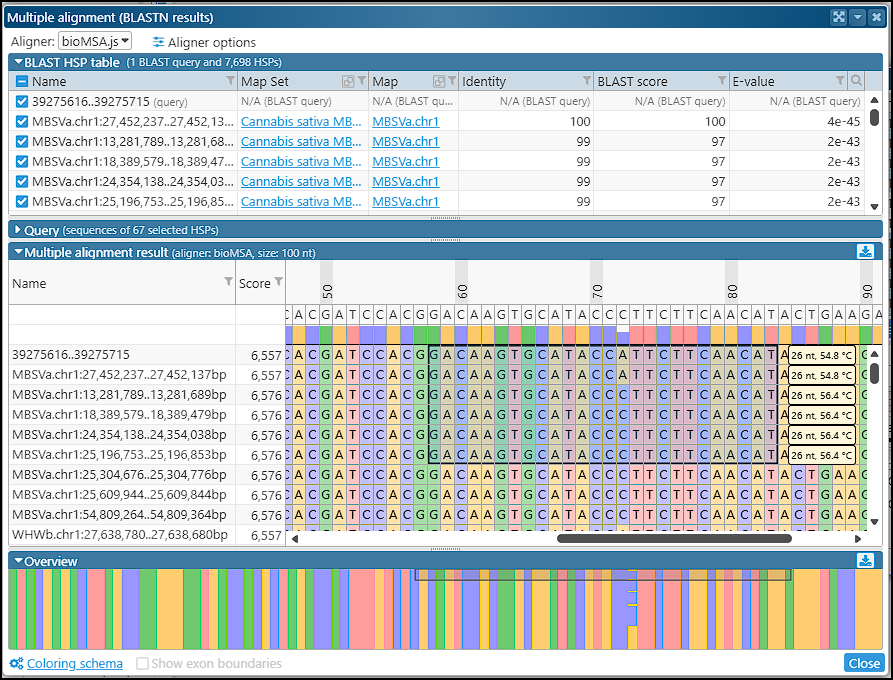
Multiple alignment for BLASTN results
We have added a helpful enhancement to the BLASTN results form: the subject sequences for each HSP can be included in the multiple alignment interface. For instance, you can select a region of interest and use its sequence as a BLASTN query to find similar sequences in other closely related genomes. From the list of results, choose the matching sequences to be used in the multiple alignment for identifying sequence variation. The query sequence will be automatically added to the list. The consensus sequence and the ratio of conservation are shown at the top of the alignment interface. This, for example, may help you with primer design when the probe sequence should target conserved regions.
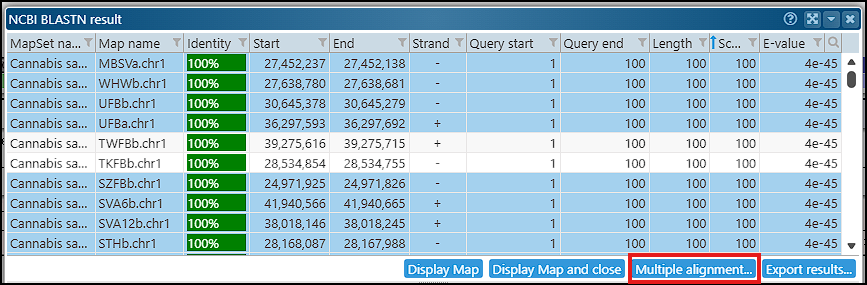
The alignment can be calculated using various algorithms (MAFFT, MUSCLE, Kalign) available in the biowasm software package, which is based on WebAssembly.