Human pangenome visualization. Phase one.
In addition to the popular versions of the Human genome, such as GRCh38 or CHM13v2, we now host full assemblies for 96 individuals from the Human pangenome. The sequences were downloaded from NCBI. Unlike the well-annotated data sets, the sequences from the pangenome, in the majority, are not assembled into chromosomes, though the contigs are quite long. To find and align matching contigs, we have extracted short sequence tags (300 bp, 50,000 bp apart) from the reference GRCh38 and mapped them onto the other assemblies. The identical tags are automatically linked by Persephone. Once a region of interest in the reference sequence has been defined, its matching region in the other genomes can be found by clicking a tag and opening its “All locations” list. Select the location of the tag on other sequences that you want to visualize and bring it to the view. Resize all maps into the same scale by clicking “Align connected features” menu item for a selected tag. Study the sequence similarity by engaging minimap2 or BLASTN.
https://web.persephonesoft.com/?bookmark=DE72A160497B428F035AE63AF3CB8ED7
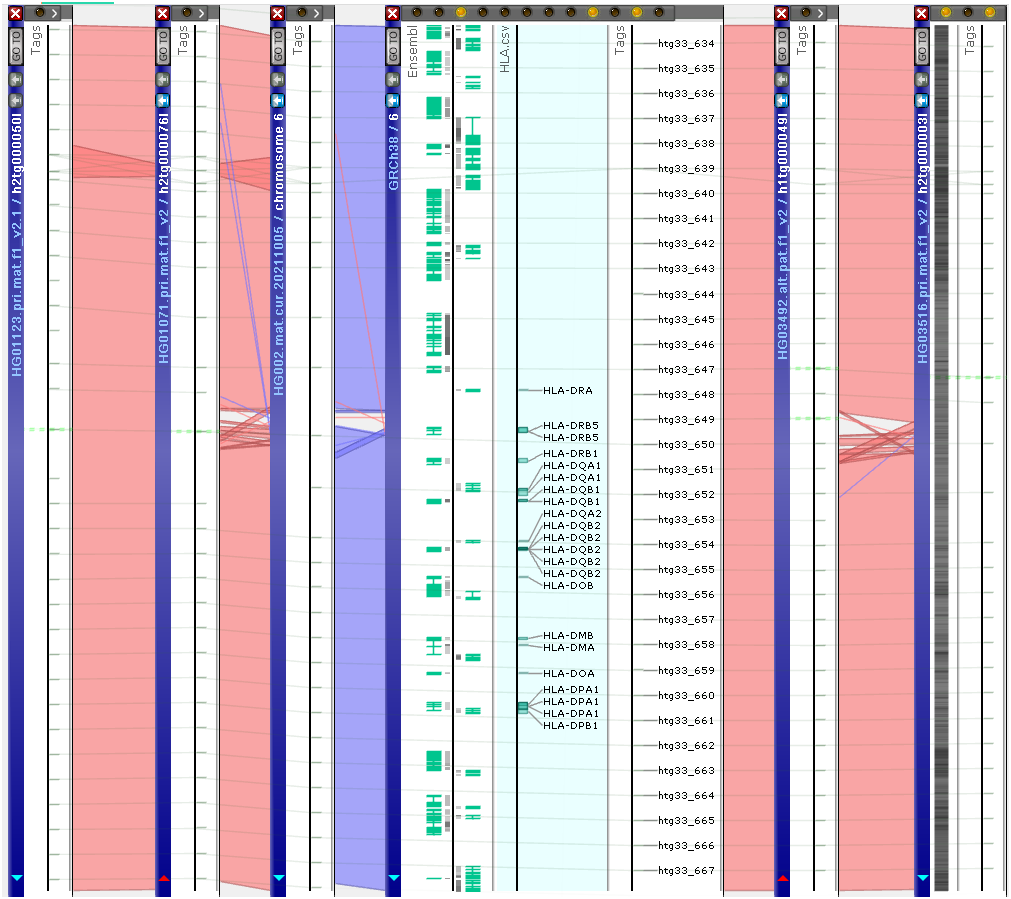
https://web.persephonesoft.com/?bookmark=C5FC10400DFF59F4767142A805802DD6

https://web.persephonesoft.com/?bookmark=2E449B7EF1FDFB361D93D05423D18DDF

This is the first phase of pangenome visualization, which allows manual analysis of the aligned sequences. The next step will be an automated exploration of the pangenome with computational discovery of the most genetically similar or distant genomes in a selected region.



