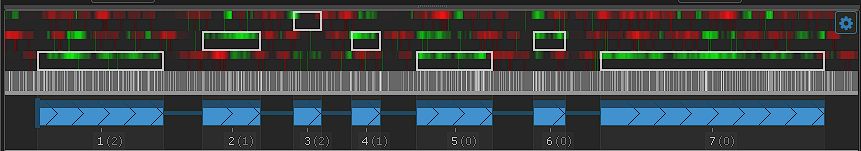
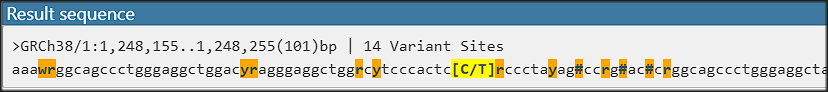
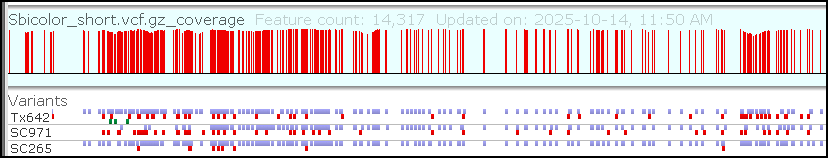
Feature update: Edit Gene Models
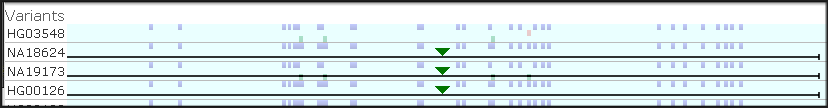
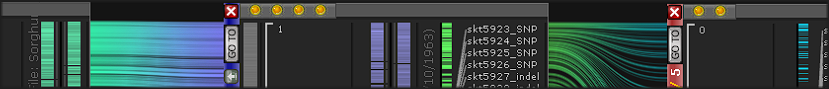
The latest addition to Persephone’s toolset is an interactive interface for editing gene models. When you encounter an issue in an existing gene model, you can now create your own corrected version of the gene structure and save it so...