A new feature: Show NGS read details
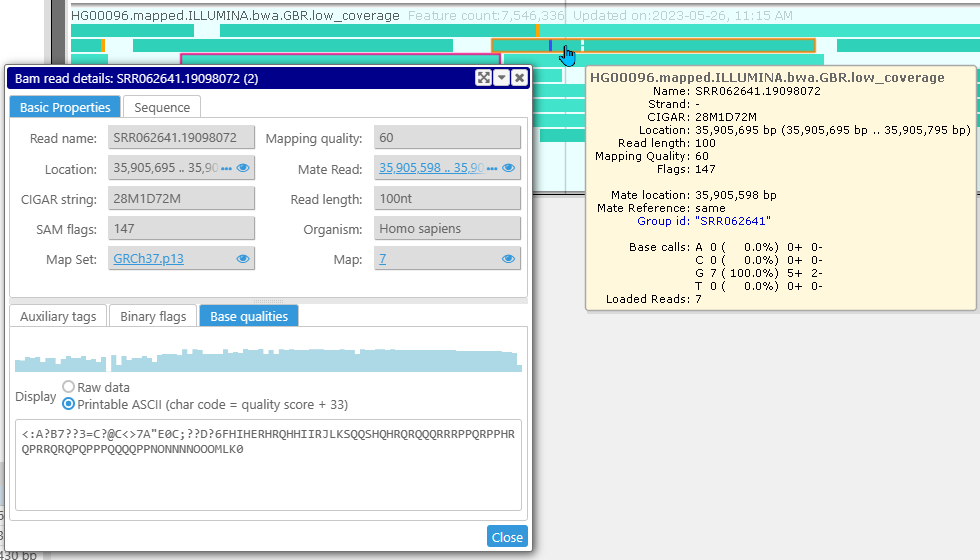
For each aligned NGS read, we have added a detailed view. If you click the read on a BAM/CRAM track, you will see the form with various properties of the read including the details of the mapping, the base qualities and a decorated read sequence.
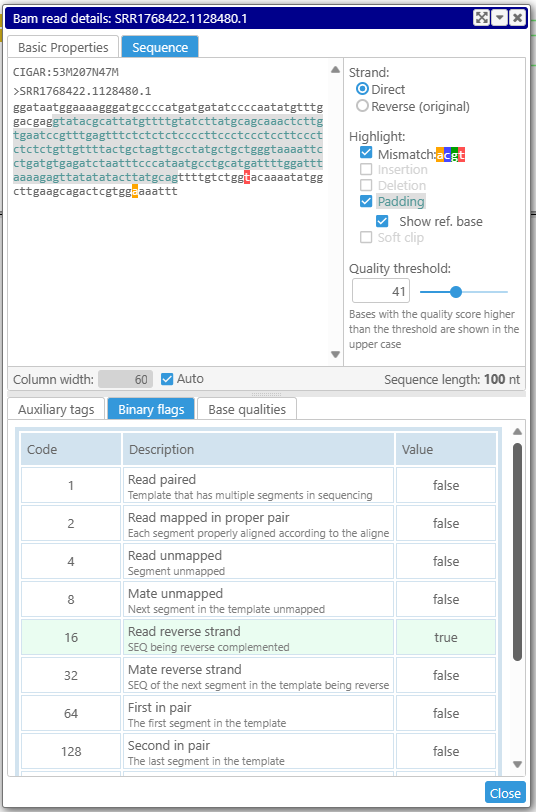
The sequence tab shows the sequence with different highlights, such as mismatches, insertions or deletions. If a read spans an intron, it is quite useful to fill the gaps with the bases from the reference, which will help verify if the intron has canonical splice sites.
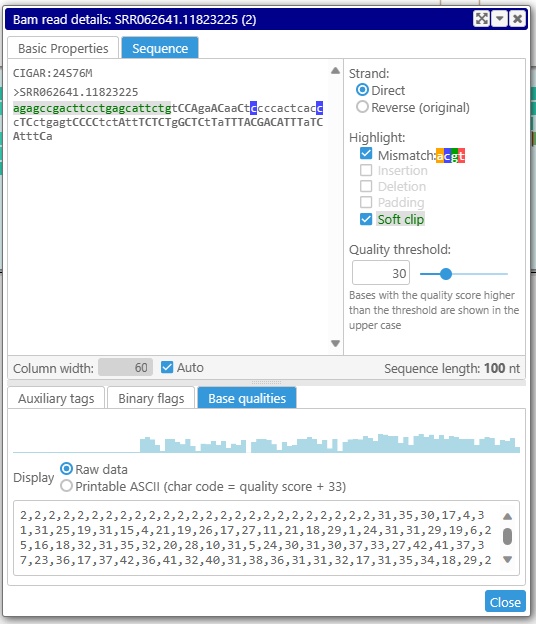
If a read is not too long (<1000 nt), the bases with the quality score higher than a threshold will be shown in the upper case. A slider can control the threshold. The quality scores are also displayed graphically in a form of a histogram.
You can hover the mouse cursor over any base in the sequence to view a tooltip listing additional information:
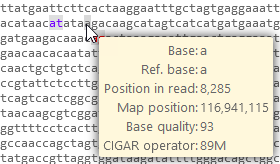
The tooltip displays the value of the base in the BAM read (if any), the value of the corresponding reference base (if any), and the exact position of the base in the read as well as on the reference map; and finally, the CIGAR operator that pertains to the highlighted position on the BAM read.
If you think that other NGS read properties should also be displayed in these forms, please let us know.



