
Feature Update: Multiple Motif Search And Restriction Sites
We have improved the motif search interface, enabling users to search for multiple motifs by entering them as a comma-separated list. Additionally, users can now select DNA motifs from the REBASE database, which contains recognition sites for restriction enzymes. To view the list of recognition sites after selecting the region for analysis, simply click the icon located inside the “Enter motif” search box.
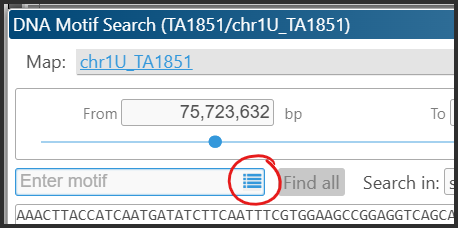
The window will be populated with the recognition sequences pulled from the REBASE database.
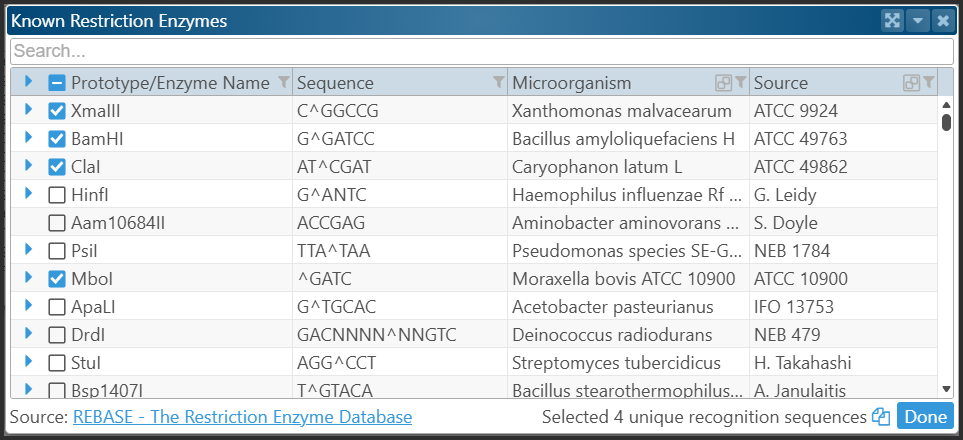
Select the enzymes you need, then click “Done.” This updates the “Enter motif” field. To search, click “Find all.” Motifs will be located in the selected region — or across the entire map, if chosen.
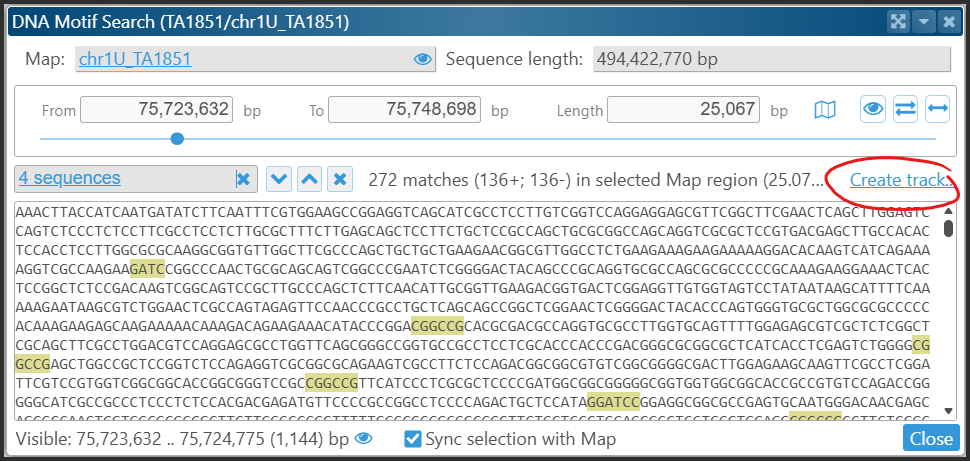
After identifying the motifs, they will be highlighted both in the text and on the Ruler track. A recent addition to the functionality is the ability to create a temporary track displaying the identified motifs. Users can choose to represent this as a marker or, if the orientation of the discovered sites is significant, as an annotation track.
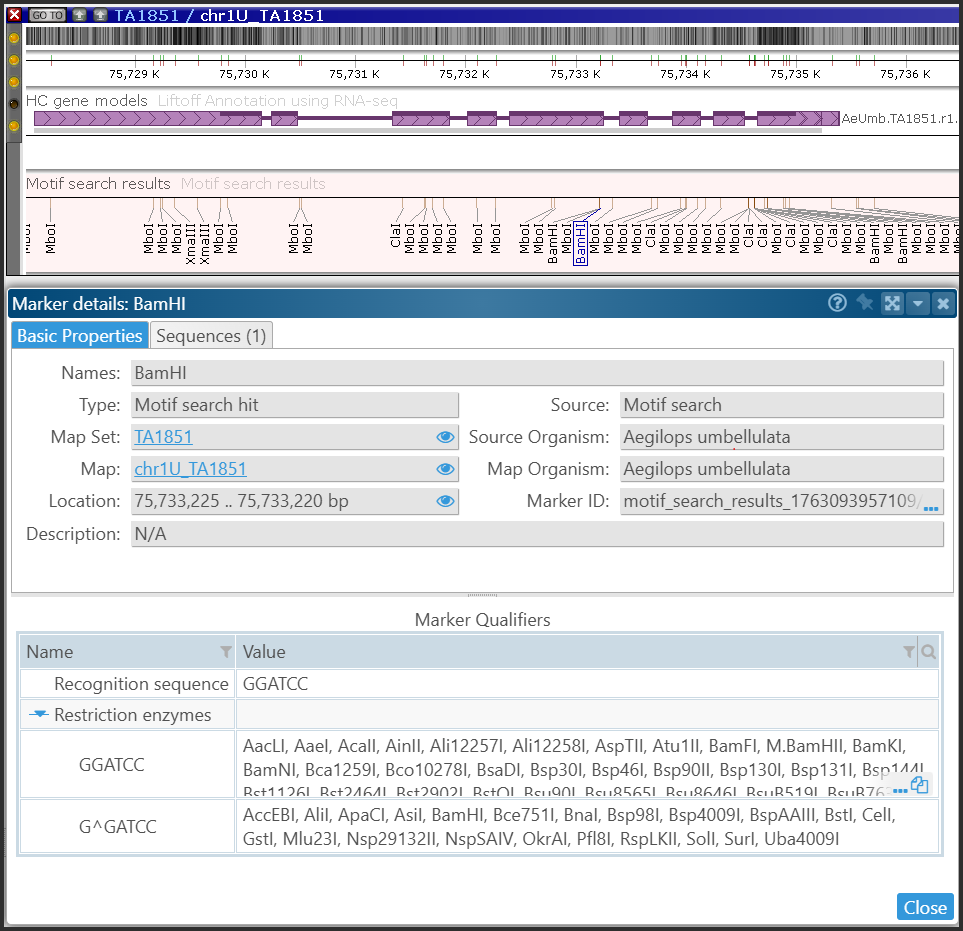
Clicking on the “marker” feature will display the properties of the recognition site. The form will also provide a list of alternative restriction enzymes that recognize the same sequence.



