Feature Update: Importing and Indexing GFF Files
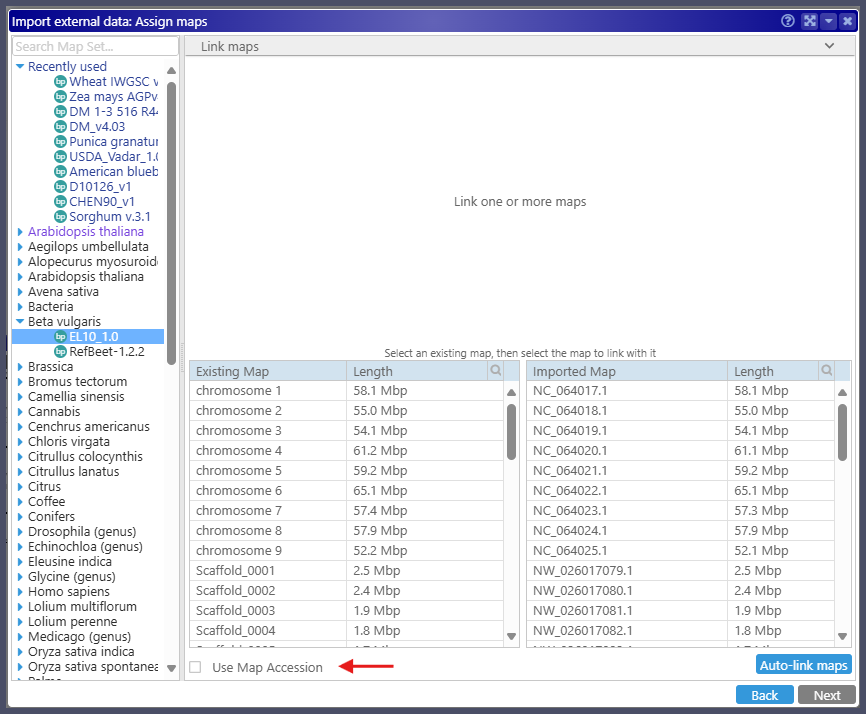
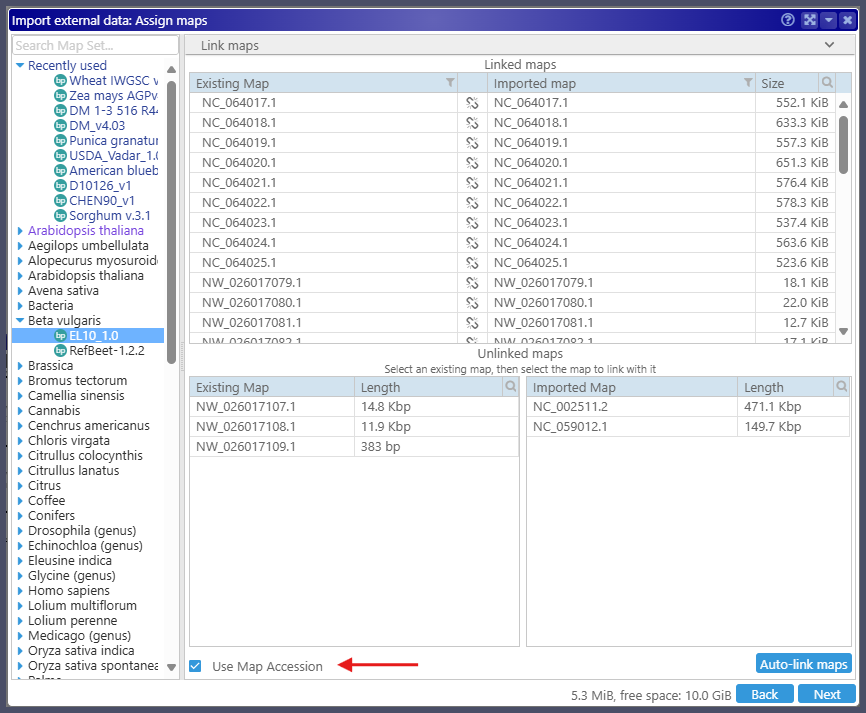
As you probably know, Persephone allows users to upload their files and view them alongside the data from the database. When importing annotation files in GFF3 format, the users must select the genome on which they want to see the new track. The import engine automatically tries to match the maps in the file to the maps in the database. A few algorithms will find the correspondence of the map names even if they are not identical. For example, the file can have names like Chr1, Chr2, etc., while the maps in the database could be named chr_1, chr_2, etc. Persephone will successfully find the matching maps. It is quite common to give the maps human-readable names. For instance, the FASTA files from Genbank have verbose headers containing the map accession at the beginning of the line and a human-readable identifier, such as ‘chromosome 1‘, embedded in the middle of the text. We can extract the map names using regular expressions when loading the data into the Persephone database. In addition, we store the obscure Genbank identifiers also parsed from the FASTA headers as map accession numbers. When asking to match the map names between the file and the database, the new feature of the import pipeline allows users to choose between map name and map accession. The maps in the Genbank GFF3 files are identified by their accession numbers. It is now easy to import such files by using the new checkbox that will allow using map accession instead of the map names.
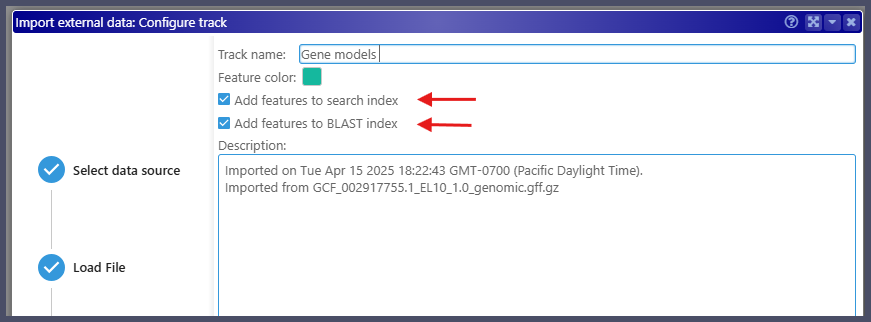
Before creating the annotation track, users can now control the data indexing. If you know in advance that the track features do not need to be searched for or are not planning to run BLASTP vs. proteins generated from the gene models, turn the corresponding checkboxes off. This may save you quite some time on the indexing process.
The status of the search indices for each imported track will be shown in the User data interface: