Feature Update: Drag & Drop VCF Files
For version 10.3.9284 and later
Until recently, the only way to visualize and analyze variants was to load VCF files into the database using our loader tool, PersephoneShell. This CPU-intensive process transformed the data, making it suitable for fast retrieval in Persephone.
We are glad to announce the latest Persephone update, which enables users to import external VCF files. These files can be referenced by URL or dragged and dropped onto the webpage. If the file is larger than 15 MB, an index file (CSI/TBI) must be provided. The data will remain in the browser’s memory until the user closes the web application. Therefore, the “local session” check box is automatically checked for the VCF files. When using the index, only relevant portions of the VCF files will be transferred on demand.
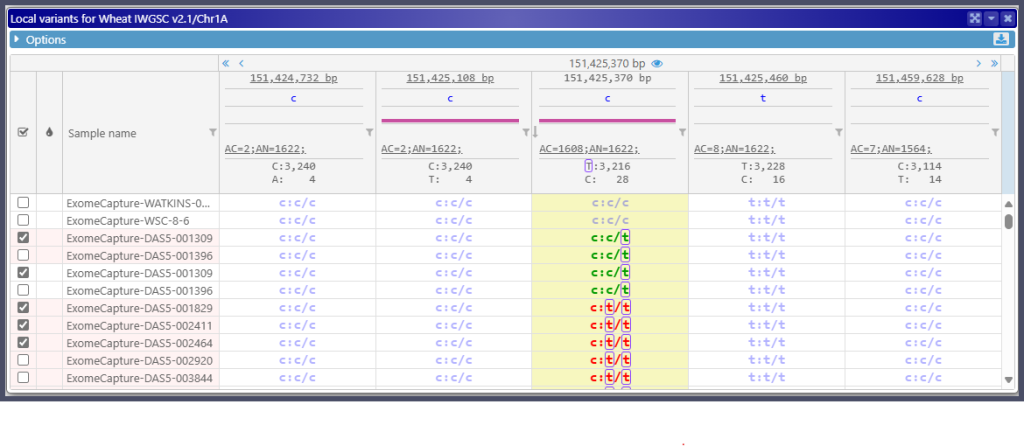
Since these files are processed directly in the browser, which usually has limited processing power, fully replicating the functionality of Persephone’s Variants module presents unique challenges. However, we’ve worked to make the experience of using external VCF files as intuitive and familiar as working with backend-loaded files. Like in other places in the Persephone application, the external file’s data is shown with a light pink background:
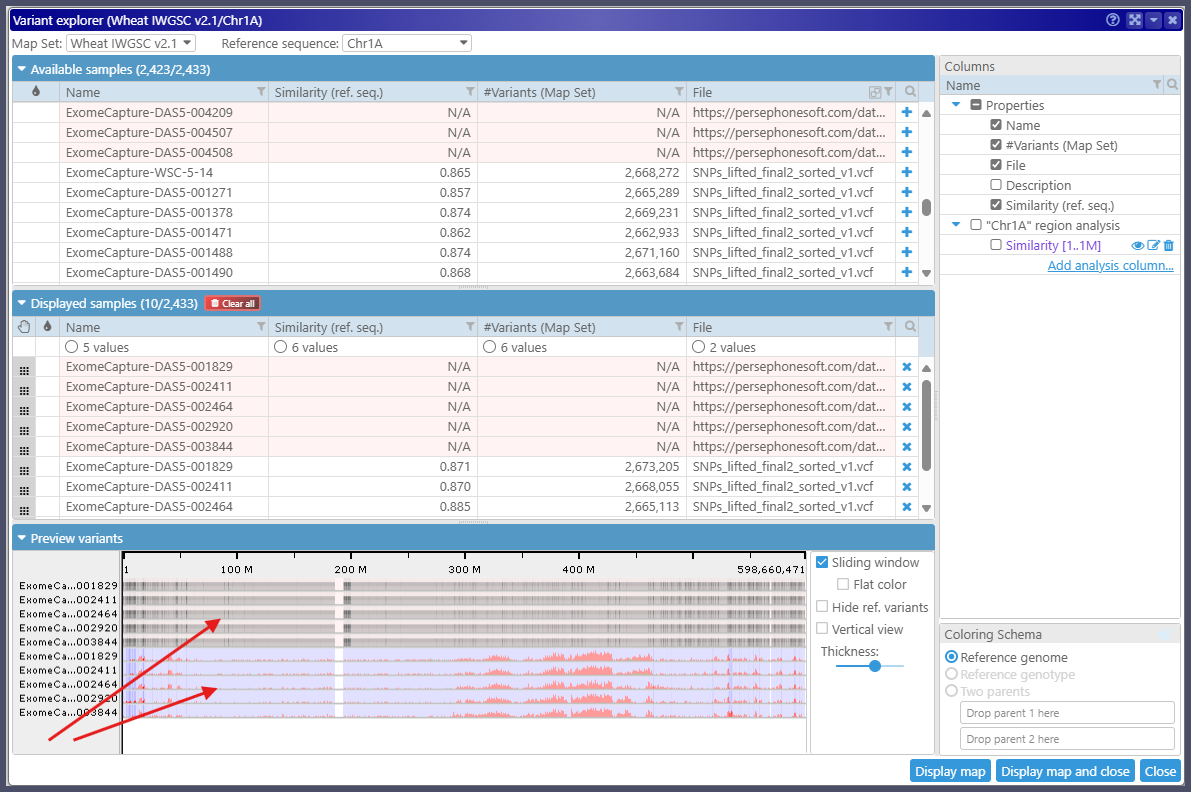
In the current implementation, we cannot afford to count all variants in the external VCF file at runtime, and accordingly, we do not calculate the similarity metric for entire chromosomes. The coverage histograms are approximated by the size of data blocks in the VCF index and, unlike the precise histograms for the variants loaded from the back end, are shown in gray. As you can see, they are somewhat similar.
Once the selected samples are displayed in the Variants track, the program treats all variants uniformly, regardless of whether they are loaded from the database or a file. Both the Local Variants and Translation Changes forms remain fully functional.