Feature Update: Multiple Protein Sequence Alignment
The new tool aligns multiple protein sequences. You can access the alignment interface via the Tools menu. In this case, the text box for the sequences will be empty, and you can populate it with the sequences you wish to align, most probably from the clipboard. Alternatively, you can open the alignment tool via the gene properties form. The Orthologs tab now has a hyperlink to show the alignment interface pre-populated with the orthologous protein sequences:
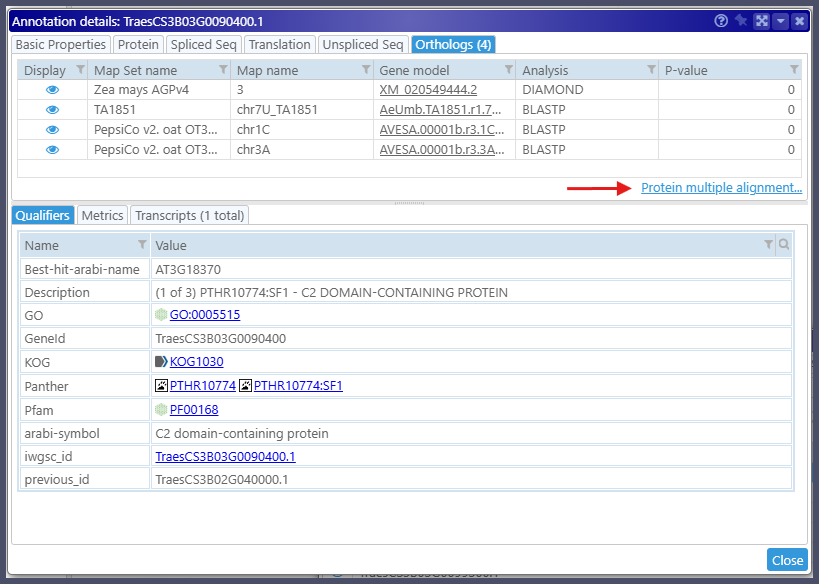
You are in control of the sequences to be aligned. By default, a group of orthologs from each map set is represented by a single protein sequence. You can add more splice variants from the orthologous genes to the alignment. The amino acid residues are painted according to a selected color schema. If you prefer, you can highlight only differences from the calculated consensus or nominate one of the proteins as a reference. Visualizing the exon boundaries in the protein sequences that, at some point, were translated from spliced transcripts will help assess the quality of the alignment.
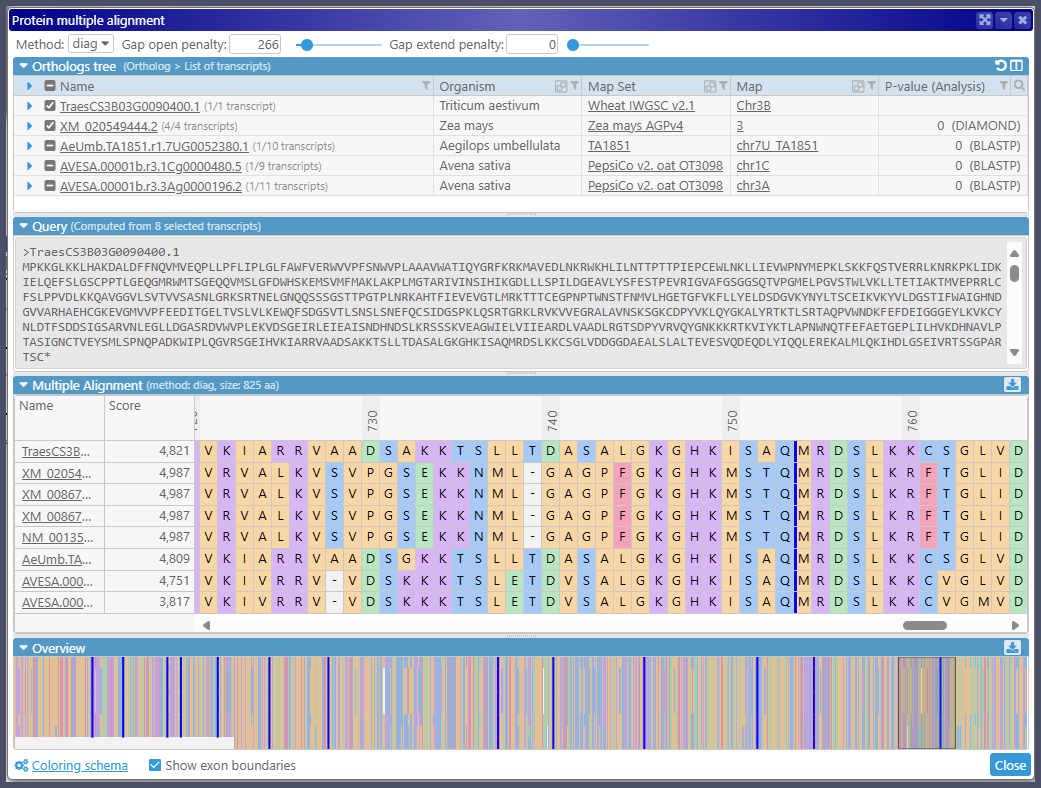
Note that the alignment can be exported in several formats.
The complete documentation of this functionality is available here.



