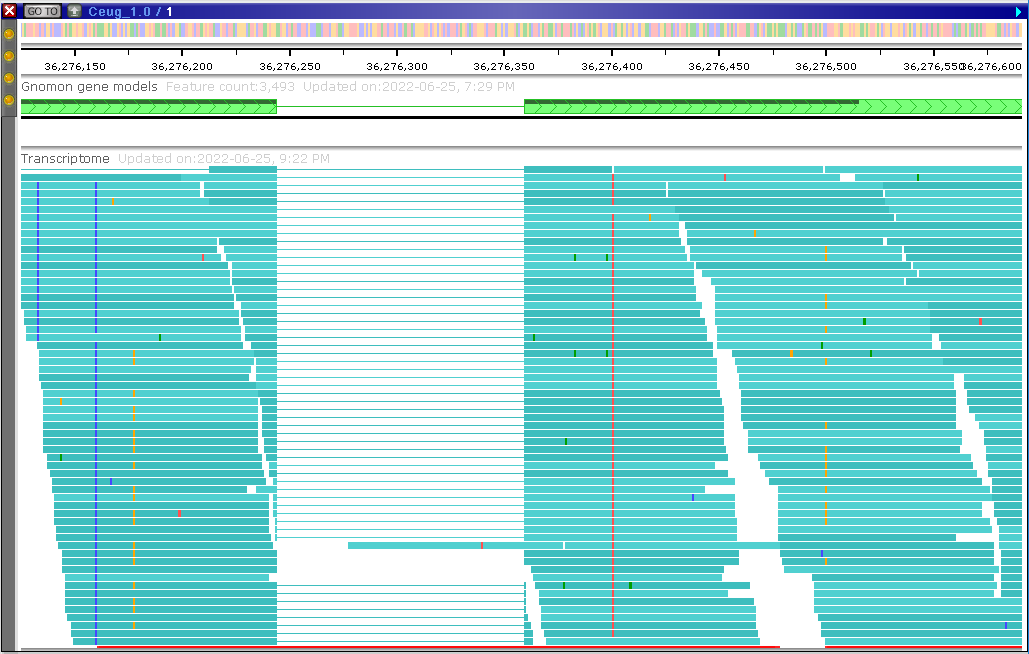
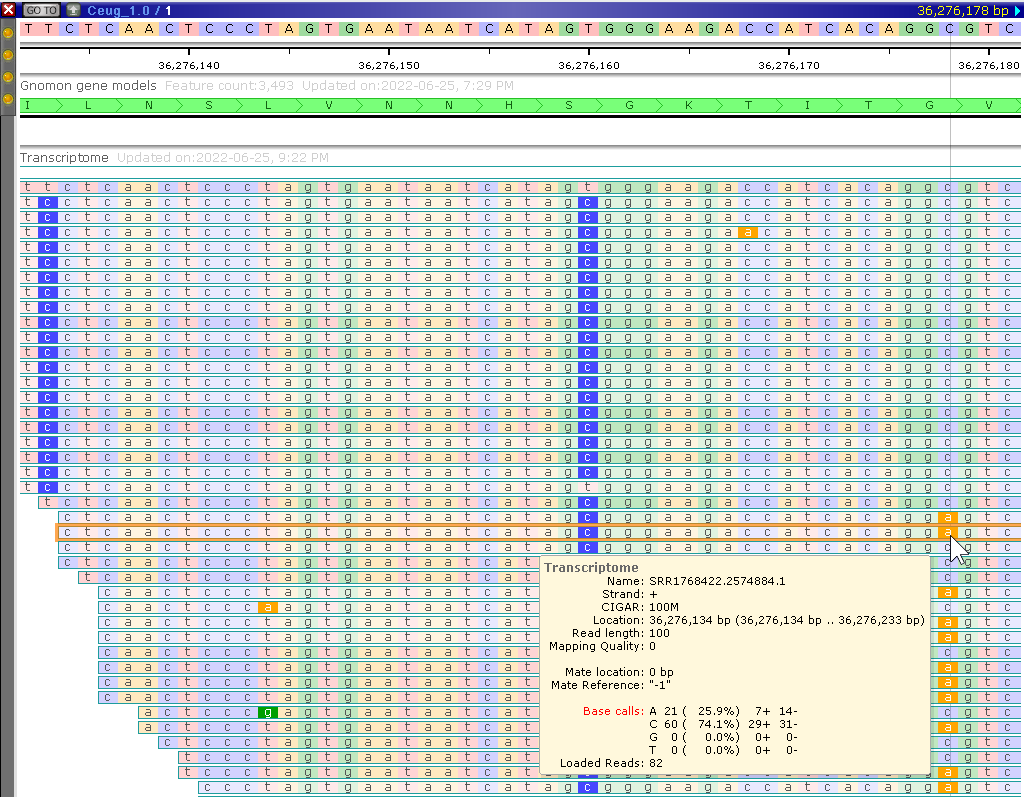
BAM tracks can be loaded permanently
The Persephone genome viewer normally visualizes the data stored in a database. The users can add their own tracks (visible to them only) by loading external files. Among the supported types of user tracks are the tracks with NGS read alignments usually in the form of BAM files. You can drag & drop files from your local computer or reference them by URL. Using the local BAM files is very efficient – the loading is almost instant, not much data travels to the server, and the browser does the decoding. This is perfect if you want to quickly look at the data. There is one drawback of this approach: the BAM track has to be temporary – in the next Persephone session, the web browser cannot open random files on your local disk (the tracks from URLs will survive).
As you probably know, the BAM files typically occupy gigabytes of disk space (we tested BAM files of 200 GB in size). So, adding such large data sets to the database can be challenging.
Luckily, we found a solution that will allow the loading of the BAM tracks to the database, making them available for all users. Our data loader PersephoneShell will load to the system only some indexing and meta-data referencing the BAM payload stored externally.
As an example of such a permanent BAM track with transcriptome alignment, take a look at the Coffea eugenioides genome in Persephone.