Persephone® Genome Browser
A platform for storing and visualizing genetic information at industrial scale.

A multi-genome browser carefully crafted using latest technologies.
Persephone® is a modern way for bioinformaticians to present genomic data to biologists
We focus on efficient storage, optimized transfer and fast rendering of genomic information. Navigating through data sets is fun. Genome is not static anymore. ``It's alive!``
We would be happy to show what a professional genome browser is capable of. Just send us an email. Thank you












and others…
A free, fully functional application is hosted on our website. The application serves data from our demo database, which is populated with numerous public genomes, such as wheat (IWGSC v2.1) or human (GRCh38). See the full list of available genomes here.
The latest news is updated on our blog.

The entire software package can be installed as a Docker container. It can run on a local machine or in the cloud
The software was designed with performance in mind, handling large datasets with ease, while keeping memory usage to a minimum.
Maps can be displayed horizontally and vertically
Persephone can be set up on premise. Our team will provide detailed instructions and will guide you through the installation process. We can also help with data loading and software integration.
Our team will work with you to develop the exact features you need.
Open local files in multiple formats, then visualize them alongside all the other data.
Persephone’s intuitive interface has been built based on suggestions from multiple researchers.
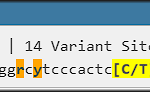
Users can run BLAST search and evaluate its results graphically, integrating them with the rest of the genomic data. With a push of a button, entire chromosomes can be aligned in real time by running minimap2.
Save displayed genomic features in various formats, including FASTA, Genbank, and CSV.
Want to host Persephone on your infrastructure and serve your own data?
Talk to us about a license
If you are affiliated with a university, you are eligible for a significantly discounted rate!
On-premise or Cloud installs
If you prefer to host the system yourself and work directly with proprietary data, we can provide a Docker image containing the complete software stack. Our team will guide you through the installation process and ensure you are fully equipped to use the system effectively.