Persephone Software at PAG33
We had a great time at the Plant and Animal Genome Conference in San Diego. Our exhibit booth and workshop drew strong interest, and it was a pleasant surprise to see several speakers highlight Persephone in their scientific presentations, which brought additional traffic our way. Visitors to the booth shared positive feedback and offered numerous ideas for future development. Many attendees specifically noted the application’s speed and intuitive design. As you may know, our web portal is free to use, and many researchers can generate meaningful results directly through our hosted instance. If a user’s preferred genomes are not yet available, they can easily upload them to their private workspace. Once a genome and its associated tracks are added, the system automatically generates a statistical summary of sequences and features, enables BLAST searches, and supports real‑time comparative genomics.
We truly enjoy interacting with our users—these conversations help us understand the direction of their research and give us the opportunity to answer questions about Persephone’s capabilities.
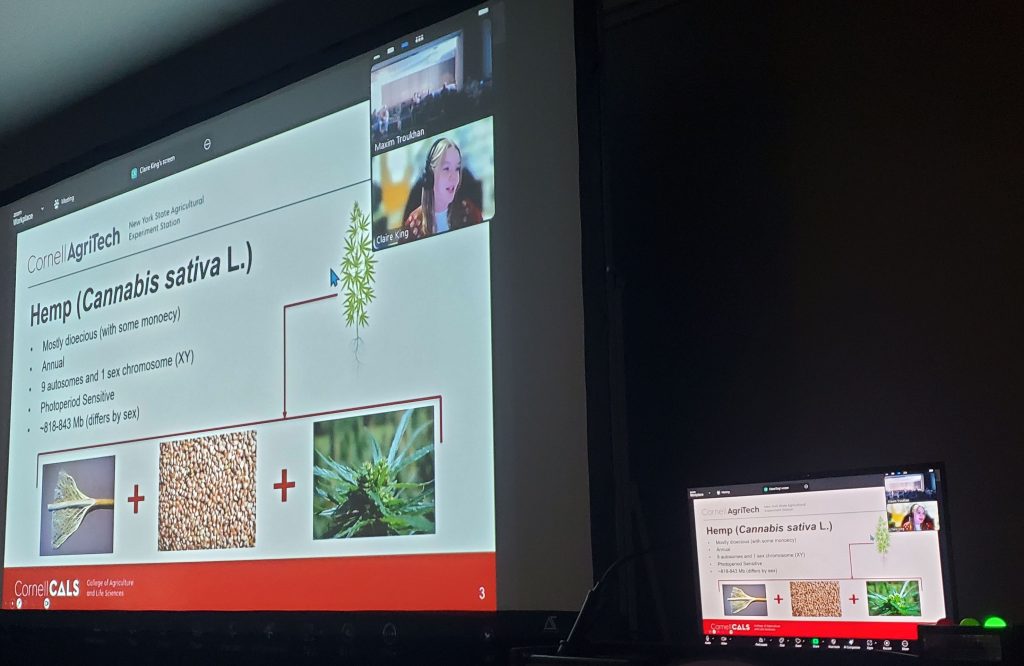
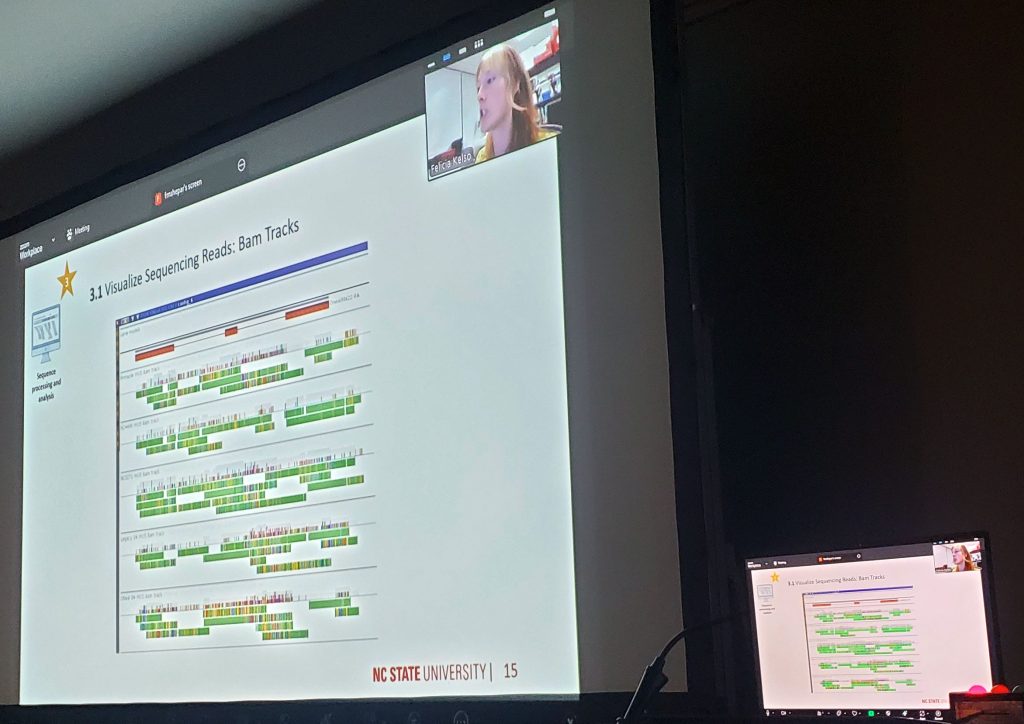
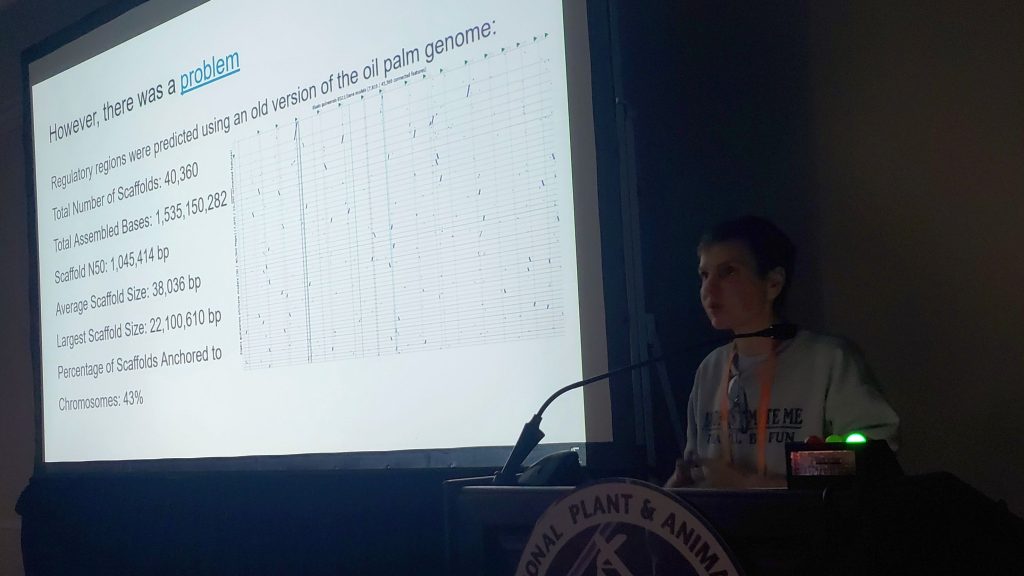
During the workshop, we reviewed current features and shared our development roadmap. One of the new directions presented by Stas Freidin is a module that will allow Persephone to read datasets directly from NCBI. This feature received an enthusiastic response; users immediately recognized its value and expressed interest in adopting it as soon as it becomes available. We also hosted several scientific talks showcasing research conducted with Persephone (Cornell’s Claire King, NCSU’s Felicia Kelso, ULV’s Tatiana Tatarinova). Some presentations were delivered via Zoom, which was well received. It was especially rewarding to see that even small features we recently introduced were already being discovered and used by researchers in real scientific workflows.
We are currently reviewing all notes and suggestions from the conference and will keep you informed as new functionality is released. Thank you for your continued feedback and support.