Persephone is available as a web application at https://web.persephonesoft.com
It is fully functional and free to use. Our portal hosts many popular genomes (about 460 genomes total). Please let us know if you don’t find your favorite organism in the list.
The application allows you to add your own datasets, which are seamlessly integrated with the system’s built‑in data. These user‑provided datasets remain private and are visible only to the account that uploaded them. Uploading data requires registration with an email address or an OpenID provider such as Google or Microsoft. Each registered user receives 5 GB of default storage.
Large BAM/CRAM files do not need to be uploaded directly. They can be accessed remotely via a URL, so only the small index file is downloaded and stored locally. This approach minimizes disk usage and prevents large files from consuming the user’s storage quota.
The full list of Persephone’s features is available here.
If the functionality available in our portal already meets your research needs, you’re welcome to continue working with it. We appreciate any feedback you can share, as it helps us refine and improve the system
If you choose to host Persephone on your own hardware and manage proprietary data within your organization, a license will be required. A local installation gives you full access to the PersephoneShell loading tool, a command‑line application capable of running multiple data‑processing and loading tasks. In addition to straightforward operations—such as importing genomic sequences or annotations—PersephoneShell can generate artificial marker tracks by extracting short sequence tags from one genome and mapping them onto others. These tracks are automatically linked through shared markers and help align maps across genomes. Map alignment can also be performed using orthologous gene pairs, which can be computed directly through another PersephoneShell command.
A personal instance offers further advantages, including the ability to add custom hyperlinks to object properties and to integrate Persephone into existing analysis pipelines. Data loading can be fully automated in batch mode; for example, Persephone can be incorporated into a gene‑discovery workflow to streamline downstream visualization and analysis.
Please let us know if you decide to host Persephone in-house.
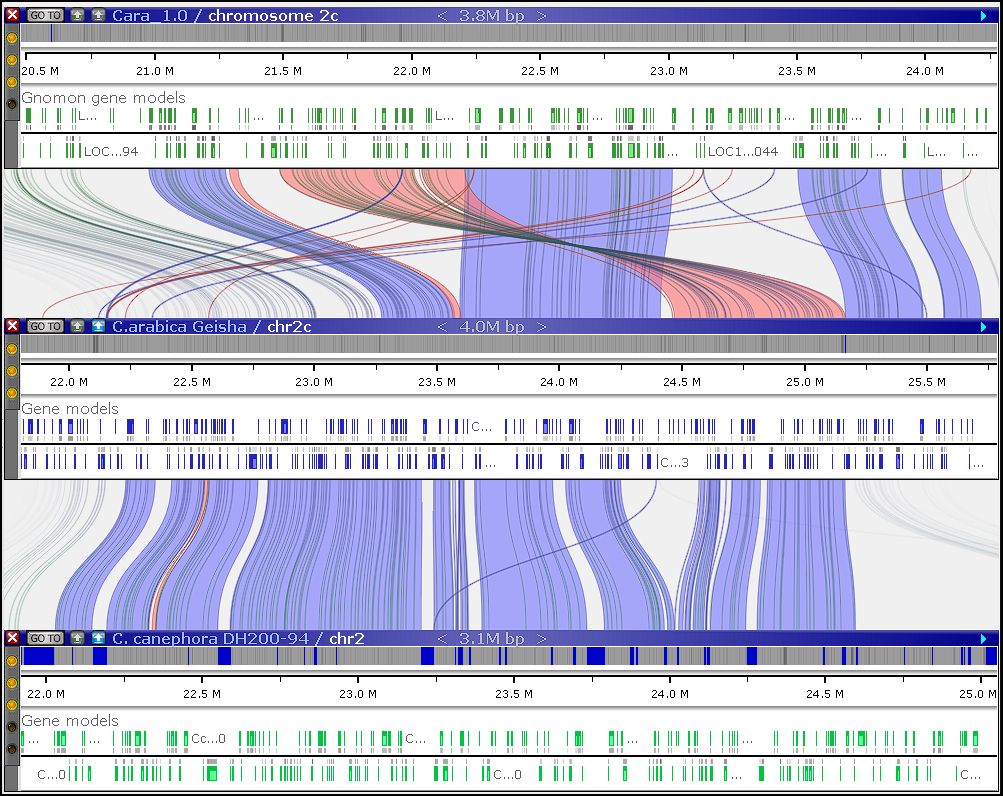
Pangenome examples:
Cannabis (97 assemblies)
Cucumber (37 assemblies)
Corn (30 assemblies)
Human (97 assemblies)
Rice (26 assemblies)
Quinoa (9 assemblies)
Solanum (34 assemblies)